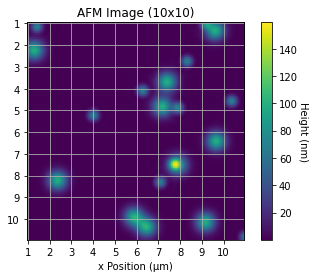
I would like to reduce the x axis from 100 μm to 10 μm and the same for the y axis. I tried setting the axis limits but want the same data to be plotted on the 10 x 10 grid that is on the 100 x 100 grid, not just a 'zoom-in'.
Here is the original code:
import numpy as np
from astropy.modeling import models
import matplotlib.pyplot as plt
z = np.zeros((100, 100))
y, x = np.mgrid[0:100, 0:100]
for _ in range(10):
g2d = models.Gaussian2D(amplitude=100,
x_mean=np.random.randint(0, 100),
y_mean=np.random.randint(0, 100),
x_stddev=3,
y_stddev=3)
z = g2d(x, y)
a2d = models.AiryDisk2D(amplitude=70,
x_0=np.random.randint(0, 100),
y_0=np.random.randint(0, 100),
radius=5)
z = a2d(x, y)
plt.imshow(z)
plt.colorbar().set_label('Height (nm)', rotation = 270)
plt.xlabel('x Position (μm)')
plt.ylabel('y Position (μm)')
plt.title('AFM Image')
plt.show()
Here is the image produced (code has a random element):
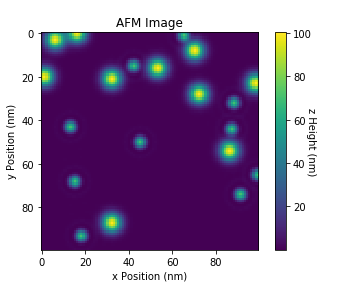
CodePudding user response:
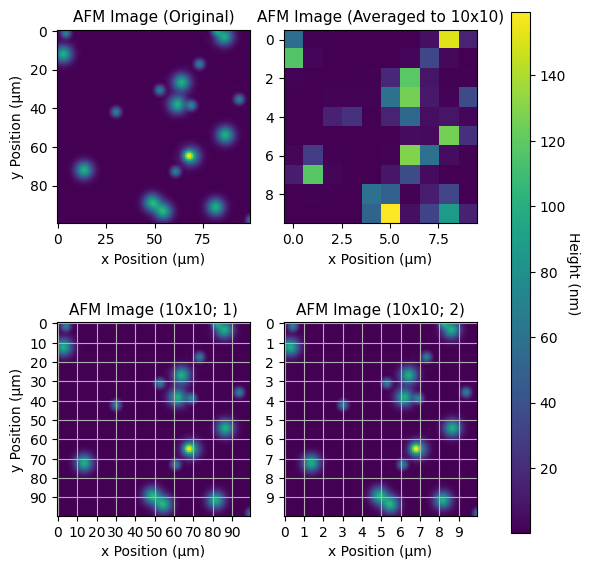
I wasn't entirely certain what you wanted- aggregation or setting new ticks, but here are a few options. See the relevant code for each plot.
If you don't want the grids on top of either image in the lower row of plots, simply remove the call to axd[…].grid()
from numpy.random import default_rng
from astropy.modeling import models
import matplotlib.pyplot as plt
from matplotlib.ticker import MultipleLocator
plt.rc('axes', titlesize=11)
rng = default_rng(0)
z = np.zeros((100, 100))
y, x = np.mgrid[0:z.shape[0], 0:z.shape[1]]
for _ in range(10):
g2d = models.Gaussian2D(amplitude=100,
x_mean=rng.uniform(0, z.shape[0]),
y_mean=rng.uniform(0, z.shape[1]),
x_stddev=3,
y_stddev=3)
z = g2d(x, y)
a2d = models.AiryDisk2D(amplitude=70,
x_0=rng.uniform(0, z.shape[0]),
y_0=rng.uniform(0, z.shape[1]),
radius=5)
z = a2d(x, y)
mosaic = [
['original', 'averaged', 'cax'],
['grid1', 'grid2', 'cax']
]
fig, axd = plt.subplot_mosaic(mosaic, figsize=(6, 6), gridspec_kw={'width_ratios': [1, 1, .1]})
# Top left plot
im = axd['original'].imshow(z)
axd['original'].set(xlabel='x Position (μm)', ylabel='y Position (μm)', title='AFM Image (Original)')
# Top right plot
m = z.reshape(10, 10, 10, 10).mean(axis=(1, 3))
axd['averaged'].imshow(m)
axd['averaged'].set(xlabel='x Position (μm)', title='AFM Image (Averaged to 10x10)')
# Lower left plot
axd['grid1'].imshow(z)
axd['grid1'].xaxis.set_major_locator(MultipleLocator(10))
axd['grid1'].yaxis.set_major_locator(MultipleLocator(10))
axd['grid1'].grid()
axd['grid1'].set(xlabel='x Position (μm)', ylabel='y Position (μm)', title='AFM Image (10x10; 1)')
# Lower right plot
axd['grid2'].imshow(z)
axd['grid2'].xaxis.set_major_locator(MultipleLocator(10))
axd['grid2'].xaxis.set_major_formatter(lambda tick, val: f'{tick // 10:g}')
axd['grid2'].yaxis.set_major_locator(MultipleLocator(10))
axd['grid2'].yaxis.set_major_formatter(lambda tick, val: f'{tick // 10:g}')
axd['grid2'].set(xlabel='x Position (μm)', title='AFM Image (10x10; 2)')
axd['grid2'].grid()
fig.colorbar(im, cax=axd['cax']).set_label('Height (nm)', rotation = 270, labelpad=10)
fig.tight_layout()
plt.show()
To create a plot with just the lower right quadrant of the above, you can use the following code:
Do note that I made a few changes to the underlying mechanics (using