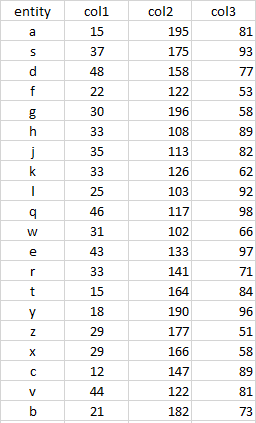
I have a dataframe as shown below:
Need to assign grouping to the entity column based on col1, col2, and col3; hence add three new columns, Group_col1, Group_col2, Group_col3
the number of groups is fixed and equals to 6 (5,4,3,2,1,0)
For each col1/col2/col3 , below calc is required to label the groups:
a. find min, max, and the mid value of the column
b. assign entity to the label '0' if the col value > mid value
c. split the remaining col values into 5 groups and assign the group labels as 5,4,3,2,1
depending upon the col values < mid value ; entity belongs to group 5 if the col values are lowest values and so on.
dput:
structure(list(entity = c("a", "s", "d", "f", "g", "h", "j",
"k", "l", "q", "w", "e", "r", "t", "y", "z", "x", "c", "v", "b"
), col1 = c(18, 27, 28, 50, 42, 18, 18, 39, 16, 49, 42, 26, 18,
38, 15, 50, 18, 10, 36, 22), col2 = c(110, 111, 159, 128, 113,
123, 128, 122, 167, 167, 113, 185, 129, 104, 180, 123, 152, 111,
117, 115), col3 = c(85, 69, 64, 96, 55, 99, 73, 55, 50, 62, 66,
87, 77, 77, 53, 60, 96, 82, 100, 55)), class = c("tbl_df", "tbl",
"data.frame"), row.names = c(NA, -20L))
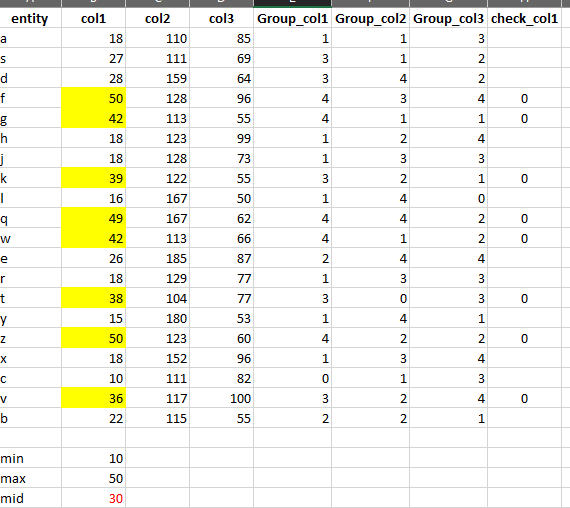
Result after running the code snippet:
CodePudding user response:
Loop across the columns that starts_with 'col', create an index to subset the values that are less than or equal to the median value ('i1'), use that subset to get the difference from the minimum value, apply cut while specifying breaks as 5, and replace the negated (i1 - greater than median value) to 0
library(dplyr)
library(tidyr)
df1 %>%
mutate(across(starts_with('col'),
~ {i1 <- .x <= median(.x)
replace(replace(rep(NA_real_, n()), i1,
as.integer(cut(-abs(.x[i1] - min(.x, na.rm = TRUE)), 5))),
!i1 ,0 )},
.names = "Group_{.col}"))
-output
# A tibble: 20 × 7
entity col1 col2 col3 Group_col1 Group_col2 Group_col3
<chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 a 18 110 85 3 4 0
2 s 27 111 69 0 4 1
3 d 28 159 64 0 0 2
4 f 50 128 96 0 0 0
5 g 42 113 55 0 3 4
6 h 18 123 99 3 1 0
7 j 18 128 73 3 0 0
8 k 39 122 55 0 1 4
9 l 16 167 50 4 0 5
10 q 49 167 62 0 0 2
11 w 42 113 66 0 3 1
12 e 26 185 87 1 0 0
13 r 18 129 77 3 0 0
14 t 38 104 77 0 5 0
15 y 15 180 53 4 0 5
16 z 50 123 60 0 1 3
17 x 18 152 96 3 0 0
18 c 10 111 82 5 4 0
19 v 36 117 100 0 2 0
20 b 22 115 55 2 3 4