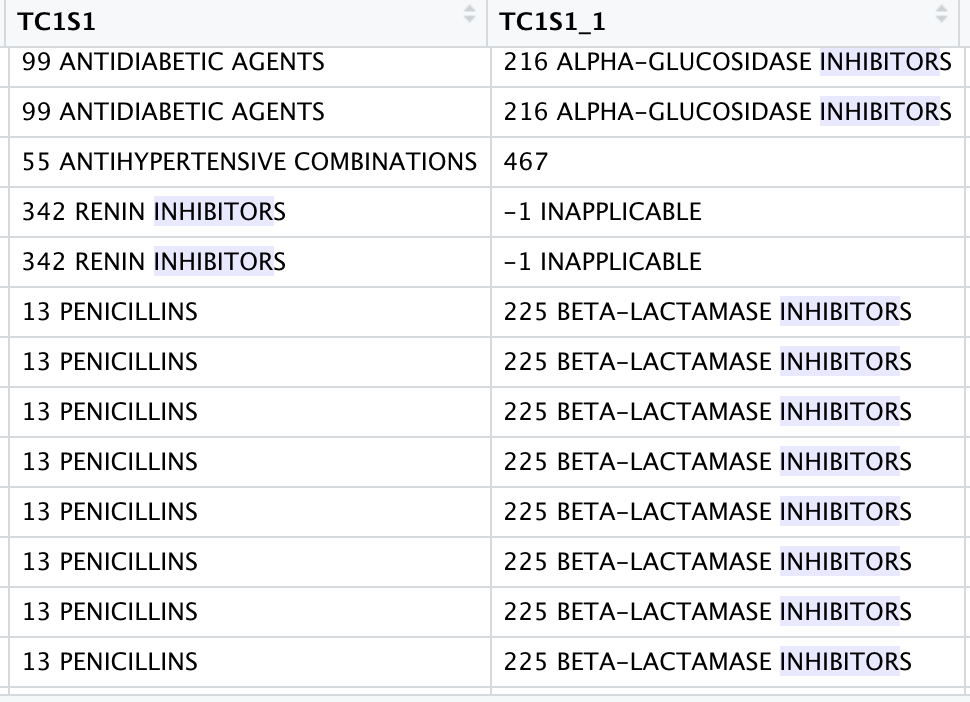
I want to subset my data frame to contain all elements that contain the word 'inhibitor'. I want to keep the entire element. For example, I'd have a new data frame with: 342 RENIN INHIBITORS, 342 RENIN INHIBITORS, 216 ALPHA-GLUCOSIDASE INHIBITORS, etc.
This doesn't work
library(dplyr)
a = data.frame(col1 = c('drug', 'drug', 'drug'),
col2 = c('drug-inhibitor', 'drug inhibitor2', 'drug'),
col3= c('drug inhibitor3', 'drug inhibitor4', 'drug'))
x <- a %>% filter(grepl('inhibitor', a[,2:3]))
In the coded example, I want a new data frame with: drug-inhibitor, drug inhibitor2, drug inhibitor3, drug inhibitor4
CodePudding user response:
We can use apply() in row mode along with grepl(), for a base R option:
a[apply(a, 1, function(r) any(grepl("inhibitor", r, fixed=TRUE))), ]
col1 col2 col3
1 drug drug-inhibitor drug inhibitor3
2 drug drug inhibitor2 drug inhibitor4
Data:
a <- data.frame(col1=c('drug', 'drug', 'drug'),
col2=c('drug-inhibitor', 'drug inhibitor2', 'drug'),
col3=c('drug inhibitor3', 'drug inhibitor4', 'drug'))
CodePudding user response:
You could also use str_detect(), e.g.
library(dplyr)
library(stringr)
a <- data.frame(
col1 = c("drug", "drug", "drug"),
col2 = c("drug-inhibitor", "drug inhibitor2", "drug"),
col3 = c("drug inhibitor3", "drug inhibitor4", "drug")
)
a %>%
filter(if_any(everything(), ~ stringr::str_detect(string = ., pattern = "inhibitor")))
Output:
col1 col2 col3
1 drug drug-inhibitor drug inhibitor3
2 drug drug inhibitor2 drug inhibitor4