I have some (legacy) code that plots a dendrogram from an n by n matrix of distances, using base R (4.1.1).
This works fine for n>=3, but fails for n=2.
numElements <- 2
data <- matrix(1, nrow = numElements, ncol = numElements)
data[1,2] <- 0
data <- (data t(data))/2
d3 <- as.dist(data)
clust3 <- hclust(d3)
plot(clust3, hang = -1)
For n=2 I get this error:
Error in graphics:::plotHclust(n1, merge, height, order(x$order), hang, : invalid dendrogram input
I would like a dendrogram with two leaves, which will show the height between just those two leaves.
Unfortunately, graphics:::plotHclust calls external C code, so I can't modify that directly. And also unfortunately, I'm trying to run this on a pre-built container on a virtual machine, so I need a base R solution without additional packages (else I'd just use ggdendro). I can catch the case of only two samples and run a separate plotting function, that is fine.
CodePudding user response:
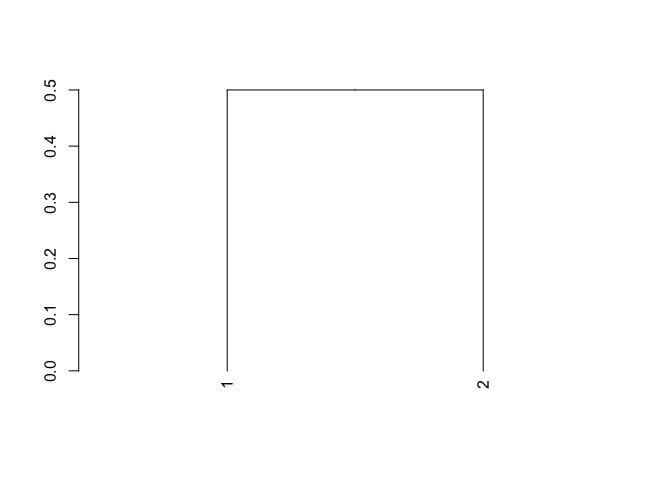
The basic dendrogram plot function can't handle a monotonic increase in the height of the gram, like described in this answer. So you could use as.dendrogram to convert it to a dendrogram object like this:
numElements <- 2
data <- matrix(1, nrow = numElements, ncol = numElements)
data[1,2] <- 0
data <- (data t(data))/2
d3 <- as.dist(data)
clust3 <- hclust(d3)
plot(as.dendrogram(clust3), hang = -1)
Created on 2022-08-19 with reprex v2.0.2
