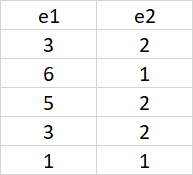
I would like to reshape the data sample below, so that to get the output like in the table. How can I reach to that? the idea is to split the column e into two columns according to the disease. Those with disease 0 in one column and those with disease 1 in the other column. thanks in advance.
structure(list(id = c(1, 2, 3, 4, 5, 6, 7, 8, 9, 10), fid = c(1,
1, 2, 2, 3, 3, 4, 4, 5, 5), disease = c(0, 1, 0, 1, 1, 0, 1, 0, 0,
1), e = c(3, 2, 6, 1, 2, 5, 2, 3, 1, 1)), class = c("tbl_df",
"tbl", "data.frame"), row.names = c(NA, -10L))
CodePudding user response:
library(tidyverse)
df %>%
pivot_wider(fid, names_from = disease, values_from = e, names_prefix = 'e') %>%
select(-fid)
e0 e1
<dbl> <dbl>
1 3 2
2 6 1
3 5 2
4 3 2
5 1 1
if you want the e1,e2 you could do:
df %>%
pivot_wider(fid, names_from = disease, values_from = e,
names_glue = 'e{disease 1}') %>%
select(-fid)
# A tibble: 5 x 2
e1 e2
<dbl> <dbl>
1 3 2
2 6 1
3 5 2
4 3 2
5 1 1
CodePudding user response:
We could use lead() combined with ìfelse statements for this:
library(dplyr)
df %>%
mutate(e2 = lead(e)) %>%
filter(row_number() %% 2 == 1) %>%
mutate(e1 = ifelse(disease==1, e2,e),
e2 = ifelse(disease==0, e2,e)) %>%
select(e1, e2)
e1 e2
<dbl> <dbl>
1 3 2
2 6 1
3 5 2
4 3 2
5 1 1