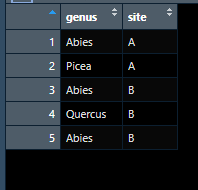
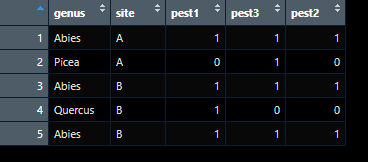
I have a list of some 500.000 trees in different sites, and am trying to find out which sites are more endangered by specific pests (n ~500). Each pest has a different host range. I have these host ranges in dataframes in a list. I am trying to use these dfs in the list as lookup tables, and will calculate fraction of suitable trees.
Example code:
#pests with their host ranges
pest1 <- as.data.frame(c("Abies", "Quercus"))
pest2 <- as.data.frame(c("Abies"))
pest3 <- as.data.frame (c("Abies", "Picea"))
pestlist <- as.list(c(pest1, pest2, pest3))
#changing this to any other kind of list would be fine too
df1 <- NULL#this will be the dataframe that would get new columns
df1$genus <- c("Abies", "Picea", "Abies", "Quercus", "Abies")
df1$site <- c("A" , "A" , "B" , "B", "B")
df1 <- as.data.frame(df1)
I tried following code, but it seems to go wrong because I don't know how to loop through lists:
library(tidyverse)
df2 <- map2(pestlist, names(df1), mutate(ifelse(df1$genus %in% pestlist , 1,0)))
For clarity, I want to go from:
to
Thanks for your time!
CodePudding user response:
If you fix up your pestlist like this:
pestlist =do.call(
rbind,lapply(seq_along(pestlist),\(i) data.frame(pest=i,genus=pestlist[[i]]))
)
then you can join df1 and a version of pestlist that has been pivoted to wide format
inner_join(
df1, pivot_wider(
mutate(pestlist,v=1),
names_from=pest,names_prefix = "pest",values_from = v,values_fill = 0
)
)
Output:
genus site pest1 pest2 pest3
1 Abies A 1 1 1
2 Picea A 0 0 1
3 Abies B 1 1 1
4 Quercus B 1 0 0
5 Abies B 1 1 1
CodePudding user response:
I would recommend using a named list of vectors instead of a list of data.frames. Then we can use map_dfc() inside mutate():
# use vectors instead of data.frames
pest1 <- c("Abies", "Quercus")
pest2 <- c("Abies")
pest3 <- c("Abies", "Picea")
df1 <- NULL#this will be the dataframe that would get new columns
df1$genus <- c("Abies", "Picea", "Abies", "Quercus", "Abies")
df1$site <- c("A" , "A" , "B" , "B", "B")
df1 <- as.data.frame(df1)
library(tidyverse)
# create named list of vectors
pestlist <- tibble::lst(pest1, pest2, pest3)
# use map_dfc
df1 %>%
mutate(map_dfc(pestlist, ~ as.integer(genus %in% .x)))
CodePudding user response:
library(data.table)
setDT(df1, key = 'genus')
pestdt = rbindlist(list(pest1, pest2, pest3), use.names = FALSE, idcol = TRUE)
setnames(pestdt, c('id', 'genus'))
pestdt = dcast(pestdt, genus ~ paste0('pest', id), value.var = 'genus', fun.aggregate = \(x) as.integer(nzchar(x)), fill = 0)
cols = paste0("pest", 1:3)
df1[, (cols) := pestdt[.SD, mget(cols)]]
#
# genus site pest1 pest2 pest3
# <char> <char> <int> <int> <int>
# 1: Abies A 1 1 1
# 2: Abies B 1 1 1
# 3: Abies B 1 1 1
# 4: Picea A 0 0 1
# 5: Quercus B 1 0 0