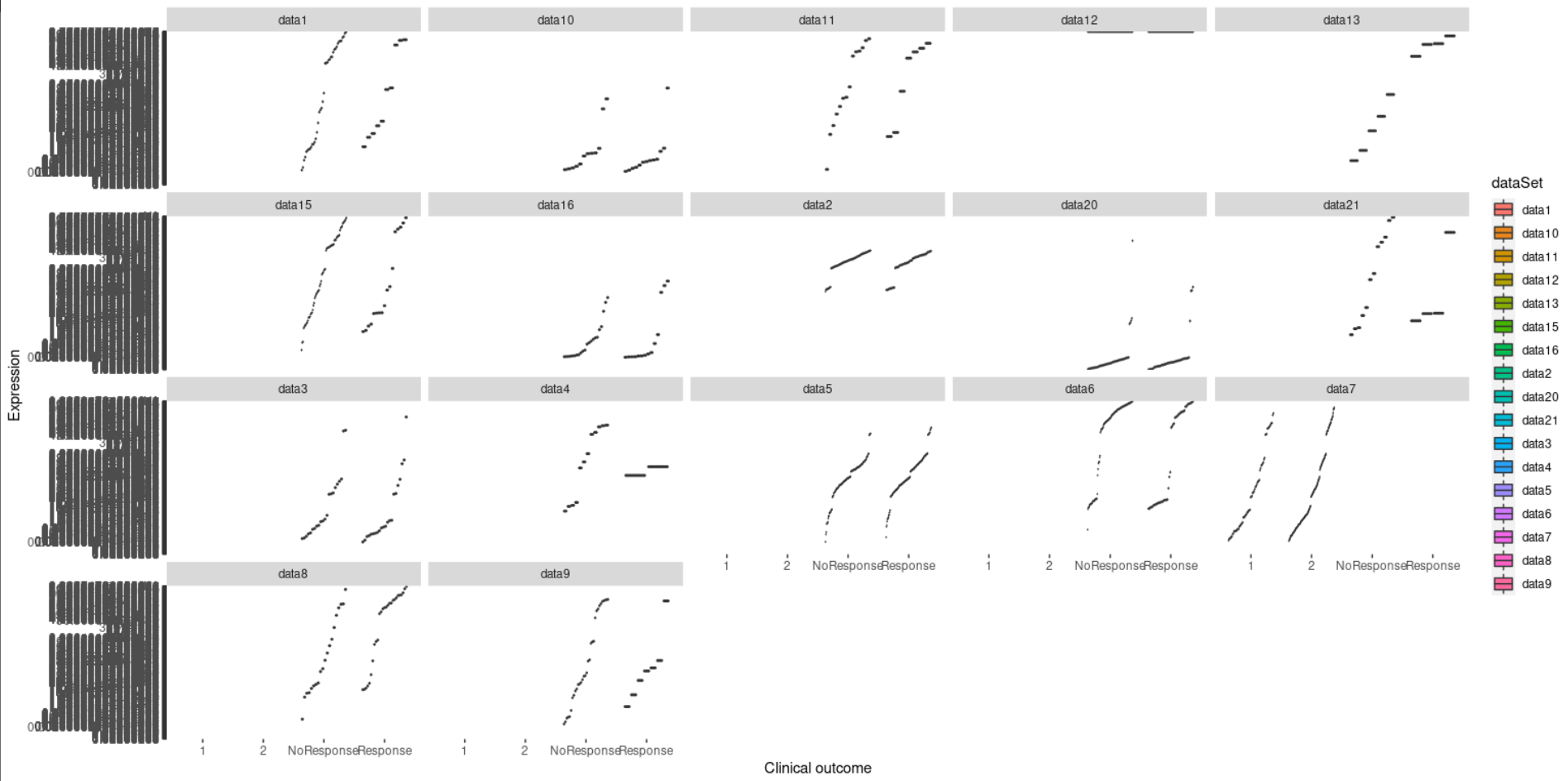
I have multiple genomic datasets about a cancer drug. The datasets are genes as rows, and samples as columns. Each dataset contains two labels, either the samples are Response or NoResponse.
Now, I want to compare the expression of a certain gene, between the Response and NoResponse groups, in each dataset individually. So the aim is that for each dataset (17 datasets overall), check if this gene is more expressed in the Response group, or the NoResponse group.
I aim to accomplish this by boxplot, using facet_wrap to indluce all boxplots in a one beautiful plot.
I'm having some trouble with that. Here is my code:
facetplot = gene_data %>% ggplot(aes(response, expr, fill = dataSet)) facet_wrap(~dataSet)
labs(x = 'Clinical outcome', y = 'Expression')
boXplots = facetplot geom_boxplot()
boXplots
I want for each boxplot in the x-axis to be the response status, and the y-axis would be the expression of the gene, and the seperation of each boxplot would be according to the dataset number.
This is a small piece of gene_data (this data is only for one gene that I chose, and it is showing samples from two datasets only, there are many more):
Each row is a sample, and you can see if the sample is Response or NoResponse, the gene expression, and to which dataset it belongs:
structure(list(expr = c("4.85031051647344", "1.53115160697977",
"12.8658881083245", "4.14991129619867", "6.38097000693584", "6.58995340312141",
"1.65500966263632", "7.12750090336409", "7.8996385796547", "7.53055682033225",
"2.22223448360773", "8.62758918096156", "3.48119701276203", "4.62943147571301",
"1.6616090122057", "2.10752212028492", "2.93218579193446", "8.14568475132326",
"13.727187056092", "3.03615586961175", "1.47868321205503", "0.486794162731451",
"2.75214758543253", "10.9209245057866", "9.81523416851324", "9.50620285828924",
"3.68894217900121", "7.99157365749216", "8.8075894831211", "2.86402639322666",
"6.80932382802606", "1.79652586144352", "4.18170107440015", "1.56494417900127",
"1.08588972884479", "2.31307162334035", "7.96791632540018", "0.286524861392658",
"5.73084282827411", "4.83366231978411", "4.51736060497412", "1.89103336082304",
"0.905882528456541", "4.24561453086179", "10.2744119421325",
"1.80851597851749", "7.85173352142177", "3.74385424752237", "1.40594503311156",
"32.95255", "32.42297", "30.17676", "29.0926", "32.06051", "31.80172",
"31.38914", "32.00126", "31.83038", "31.97847", "28.4888", "30.99362",
"29.11862", "30.76407", "31.59104", "30.68113", "31.96988", "29.31726",
"30.98914", "29.66506", "29.89776", "31.35337", "29.90499", "32.16743",
"31.22215", "32.12123", "31.78393", "30.11914", "29.54727", "31.75156",
"30.99448", "31.48003", "32.42661", "33.24433", "32.11272", "30.89568",
"28.33333", "30.87174", "32.07811", "31.05286", "30.6044", "31.62753",
"32.33149", "29.67083", "31.75701", "29.73103", "31.03143", "30.83465",
"27.53271", "33.15345", "31.47515"), response = c("NoResponse",
"NoResponse", "Response", "NoResponse", "NoResponse", "NoResponse",
"NoResponse", "Response", "NoResponse", "NoResponse", "NoResponse",
"NoResponse", "NoResponse", "NoResponse", "Response", "Response",
"NoResponse", "Response", "NoResponse", "NoResponse", "NoResponse",
"NoResponse", "NoResponse", "Response", "NoResponse", "NoResponse",
"Response", "Response", "NoResponse", "NoResponse", "NoResponse",
"NoResponse", "NoResponse", "NoResponse", "NoResponse", "Response",
"NoResponse", "NoResponse", "NoResponse", "NoResponse", "NoResponse",
"NoResponse", "NoResponse", "NoResponse", "NoResponse", "NoResponse",
"NoResponse", "Response", "NoResponse", "Response", "NoResponse",
"NoResponse", "NoResponse", "Response", "Response", "NoResponse",
"NoResponse", "Response", "Response", "NoResponse", "Response",
"NoResponse", "Response", "NoResponse", "NoResponse", "Response",
"Response", "Response", "NoResponse", "NoResponse", "NoResponse",
"NoResponse", "NoResponse", "NoResponse", "NoResponse", "Response",
"Response", "Response", "NoResponse", "NoResponse", "NoResponse",
"NoResponse", "Response", "NoResponse", "NoResponse", "Response",
"Response", "Response", "Response", "NoResponse", "NoResponse",
"NoResponse", "NoResponse", "NoResponse", "Response", "Response",
"NoResponse", "Response", "NoResponse", "NoResponse"), dataSet = c("data1",
"data1", "data1", "data1", "data1", "data1", "data1", "data1",
"data1", "data1", "data1", "data1", "data1", "data1", "data1",
"data1", "data1", "data1", "data1", "data1", "data1", "data1",
"data1", "data1", "data1", "data1", "data1", "data1", "data1",
"data1", "data1", "data1", "data1", "data1", "data1", "data1",
"data1", "data1", "data1", "data1", "data1", "data1", "data1",
"data1", "data1", "data1", "data1", "data1", "data1", "data2",
"data2", "data2", "data2", "data2", "data2", "data2", "data2",
"data2", "data2", "data2", "data2", "data2", "data2", "data2",
"data2", "data2", "data2", "data2", "data2", "data2", "data2",
"data2", "data2", "data2", "data2", "data2", "data2", "data2",
"data2", "data2", "data2", "data2", "data2", "data2", "data2",
"data2", "data2", "data2", "data2", "data2", "data2", "data2",
"data2", "data2", "data2", "data2", "data2", "data2", "data2",
"data2")), row.names = c(NA, 100L), class = "data.frame")
My code is producing this weird output:
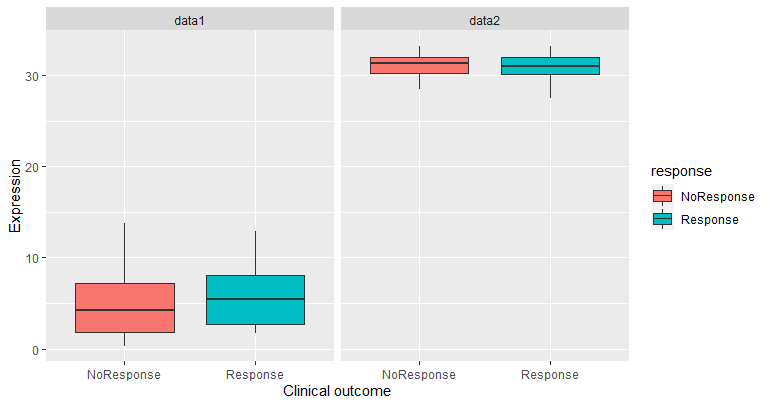
Wanted output - something like this, where the blue boxes are for Response and the red are for NoResponse:
CodePudding user response:
As per @neilws comment, the expression values are strings that need to be changed to numeric:
gene_data$expr = as.numeric(as.character(gene_data$expr))
And to match the example given you would need to change fill to response
ggplot(gene_data,aes(response, expr, fill = response))
geom_boxplot()
facet_wrap(~dataSet)
labs(x = 'Clinical outcome', y = 'Expression')