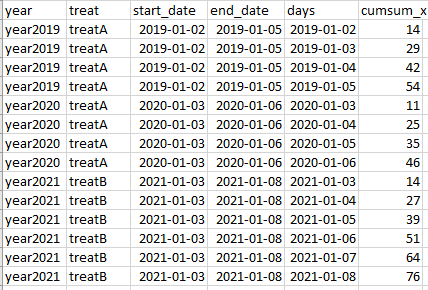
I have this data set:
library(dplyr)
library(lubridate)
data_a <- read.csv(text = "
date,variable_x
2019-01-01,13
2019-01-02,14
2019-01-03,15
2019-01-04,13
2019-01-05,12
2019-01-06,11
2019-01-07,11
2019-01-08,11
2020-01-01,12
2020-01-02,12
2020-01-03,11
2020-01-04,13
2020-01-05,10
2020-01-06,11
2020-01-07,12
2020-01-08,10
2021-01-01,12
2021-01-02,12
2021-01-03,14
2021-01-04,14
2021-01-05,12
2021-01-06,13
2021-01-07,13
2021-01-08,11
") %>%
mutate(date = as.Date(date, format = "%Y-%m-%d"),
variable_x = as.numeric(variable_x))
Also, this additional dataset sets the limits of the dates I am interested in:
data_b <- read.csv(text = "
year,treat,start_date,end_date
year2019,treatA,2019-01-02,2019-01-05
year2020,treatA,2020-01-03,2020-01-06
year2021,treatB,2021-01-03,2021-01-08
") %>%
mutate(start_date = as.Date(start_date, format = "%Y-%m-%d"),
end_date = as.Date(end_date, format = "%Y-%m-%d"))
The outcome I am looking for is this:
where I first expand the days between the start_date and end_date for each combination of year and treat, and then I calculate the cumsum of the variable_x. After searching around I approximated this solution that is not working:
outcome <- data_b %>%
group_by(year, treat) %>%
mutate(id = 1:nrow(.)) %>%
rowwise() %>%
do(data.frame(id=.$id, days=seq(.$start_date,.$end_date,by="days"))) %>%
mutate(cumsum_x = cumsum(data_a$variable_x[data_a$date %within% interval(start_date, end_date)]))
This is the error I am getting:
Error in `mutate()`:
! Problem while computing `id = 1:nrow(.)`.
x `id` must be size 1, not 3.
ℹ The error occurred in group 1: year = "year2019", treat = "treatA".
Run `rlang::last_error()` to see where the error occurred.
Any help will be really appreciated.
CodePudding user response:
The ivs package can be used for this. It is a package for working with intervals of data.
library(dplyr)
library(tidyr)
library(ivs)
data_a <- read.csv(text = "
date,variable_x
2019-01-01,13
2019-01-02,14
2019-01-03,15
2019-01-04,13
2019-01-05,12
2019-01-06,11
2019-01-07,11
2019-01-08,11
2020-01-01,12
2020-01-02,12
2020-01-03,11
2020-01-04,13
2020-01-05,10
2020-01-06,11
2020-01-07,12
2020-01-08,10
2021-01-01,12
2021-01-02,12
2021-01-03,14
2021-01-04,14
2021-01-05,12
2021-01-06,13
2021-01-07,13
2021-01-08,11
") %>%
mutate(date = as.Date(date, format = "%Y-%m-%d"),
variable_x = as.numeric(variable_x))
data_b <- read.csv(text = "
year,treat,start_date,end_date
year2019,treatA,2019-01-02,2019-01-05
year2020,treatA,2020-01-03,2020-01-06
year2021,treatB,2021-01-03,2021-01-08
") %>%
mutate(start_date = as.Date(start_date, format = "%Y-%m-%d"),
end_date = as.Date(end_date, format = "%Y-%m-%d"))
# We do ` 1L` to your end dates because ivs uses right-open intervals
data_b <- data_b %>%
as_tibble() %>%
mutate(end_date = end_date 1L) %>%
mutate(range = iv(start_date, end_date), .keep = "unused")
data_b
#> # A tibble: 3 × 3
#> year treat range
#> <chr> <chr> <iv<date>>
#> 1 year2019 treatA [2019-01-02, 2019-01-06)
#> 2 year2020 treatA [2020-01-03, 2020-01-07)
#> 3 year2021 treatB [2021-01-03, 2021-01-09)
# Find all instances of where `date_a$date` is between the range defined
# by `data_b$range`
locs <- iv_locate_between(data_a$date, data_b$range, no_match = "drop")
locs
#> needles haystack
#> 1 2 1
#> 2 3 1
#> 3 4 1
#> 4 5 1
#> 5 11 2
#> 6 12 2
#> 7 13 2
#> 8 14 2
#> 9 19 3
#> 10 20 3
#> 11 21 3
#> 12 22 3
#> 13 23 3
#> 14 24 3
# Use the overlap locations from above to join the two data frames
joined <- iv_align(data_a, data_b, locations = locs) %>%
as_tibble() %>%
unpack(c(needles, haystack))
# Group by `range` and compute the cumulative sum
joined %>%
group_by(range) %>%
mutate(cumsum_x = cumsum(variable_x)) %>%
ungroup()
#> # A tibble: 14 × 6
#> date variable_x year treat range cumsum_x
#> <date> <dbl> <chr> <chr> <iv<date>> <dbl>
#> 1 2019-01-02 14 year2019 treatA [2019-01-02, 2019-01-06) 14
#> 2 2019-01-03 15 year2019 treatA [2019-01-02, 2019-01-06) 29
#> 3 2019-01-04 13 year2019 treatA [2019-01-02, 2019-01-06) 42
#> 4 2019-01-05 12 year2019 treatA [2019-01-02, 2019-01-06) 54
#> 5 2020-01-03 11 year2020 treatA [2020-01-03, 2020-01-07) 11
#> 6 2020-01-04 13 year2020 treatA [2020-01-03, 2020-01-07) 24
#> 7 2020-01-05 10 year2020 treatA [2020-01-03, 2020-01-07) 34
#> 8 2020-01-06 11 year2020 treatA [2020-01-03, 2020-01-07) 45
#> 9 2021-01-03 14 year2021 treatB [2021-01-03, 2021-01-09) 14
#> 10 2021-01-04 14 year2021 treatB [2021-01-03, 2021-01-09) 28
#> 11 2021-01-05 12 year2021 treatB [2021-01-03, 2021-01-09) 40
#> 12 2021-01-06 13 year2021 treatB [2021-01-03, 2021-01-09) 53
#> 13 2021-01-07 13 year2021 treatB [2021-01-03, 2021-01-09) 66
#> 14 2021-01-08 11 year2021 treatB [2021-01-03, 2021-01-09) 77
CodePudding user response:
You can do this with a non-equi join in data.table:
# load library
library(data.table)
# set your tables to be data.table
setDT(data_a); setDT(data_b)
# non-equi join, cumsum by year/treat, and select columns
data_a[, d:=date][data_b, on=.(d>=start_date, d<=end_date)][
, .(start_date=d, end_date=d.1, days=date, cumsum_x= cumsum(variable_x)), .(year,treat)]
Output:
year treat start_date end_date days cumsum_x
1: year2019 treatA 2019-01-02 2019-01-05 2019-01-02 14
2: year2019 treatA 2019-01-02 2019-01-05 2019-01-03 29
3: year2019 treatA 2019-01-02 2019-01-05 2019-01-04 42
4: year2019 treatA 2019-01-02 2019-01-05 2019-01-05 54
5: year2020 treatA 2020-01-03 2020-01-06 2020-01-03 11
6: year2020 treatA 2020-01-03 2020-01-06 2020-01-04 24
7: year2020 treatA 2020-01-03 2020-01-06 2020-01-05 34
8: year2020 treatA 2020-01-03 2020-01-06 2020-01-06 45
9: year2021 treatB 2021-01-03 2021-01-08 2021-01-03 14
10: year2021 treatB 2021-01-03 2021-01-08 2021-01-04 28
11: year2021 treatB 2021-01-03 2021-01-08 2021-01-05 40
12: year2021 treatB 2021-01-03 2021-01-08 2021-01-06 53
13: year2021 treatB 2021-01-03 2021-01-08 2021-01-07 66
14: year2021 treatB 2021-01-03 2021-01-08 2021-01-08 77
Another option is to use data.table::foverlaps(). For this, to work, you need to set the key on data_b, and (like above) add a second date variable to data_a (because, while the start and end can be equal to each other in foverlaps, they can't be the same column):
setDT(data_a);setDT(data_b)
data_a[, d:=date]
setkey(data_b, start_date, end_date)
foverlaps(
data_a,data_b,
by.x = c("date", "d"), by.y=c("start_date", "end_date"),
nomatch=0)[,cumsum_x:=cumsum(variable_x), .(year, treat)][
, `:=`(variable_x=NULL, d=NULL)][]
CodePudding user response:
Here's a more tidyverse friendly solution:
create a list of dates from your data_a df:
library(tidyverse)
dates_ls <- mapply(seq.Date, data_b$start_date, data_b$end_date, by = 1) %>%
map(enframe, name = NULL) %>%
bind_rows(.id = "index") # index for merging later
... add an index to data_b:
data_bi <- data_b %>%
mutate(index = row_number())
... merge to get in long format for all the dates inbetween date ranges:
data_b_merge <- merge(dates_ls, data_bi, by = "index")
... and finally merge back to data_a and calculate the cumsum by the groups you mentioned:
data_merge <- merge(data_a, data_b_merge, by.x = "date", by.y = "value") %>%
group_by(year, treat) %>%
mutate(cumsum_x = cumsum(variable_x)) %>%
ungroup() %>%
select(year, treat, start_date, end_date, days = date, cumsum_x)
to get:
# A tibble: 14 × 6
year treat start_date end_date days cumsum_x
<chr> <chr> <date> <date> <date> <dbl>
1 year2019 treatA 2019-01-02 2019-01-05 2019-01-02 14
2 year2019 treatA 2019-01-02 2019-01-05 2019-01-03 29
3 year2019 treatA 2019-01-02 2019-01-05 2019-01-04 42
4 year2019 treatA 2019-01-02 2019-01-05 2019-01-05 54
5 year2020 treatA 2020-01-03 2020-01-06 2020-01-03 11
6 year2020 treatA 2020-01-03 2020-01-06 2020-01-04 24
7 year2020 treatA 2020-01-03 2020-01-06 2020-01-05 34
8 year2020 treatA 2020-01-03 2020-01-06 2020-01-06 45
9 year2021 treatB 2021-01-03 2021-01-08 2021-01-03 14
10 year2021 treatB 2021-01-03 2021-01-08 2021-01-04 28
11 year2021 treatB 2021-01-03 2021-01-08 2021-01-05 40
12 year2021 treatB 2021-01-03 2021-01-08 2021-01-06 53
13 year2021 treatB 2021-01-03 2021-01-08 2021-01-07 66
14 year2021 treatB 2021-01-03 2021-01-08 2021-01-08 77
CodePudding user response:
I thought you could merge data_a with an outcome that was just built with the do.call(data.frame( ...) that you had made. The rowwise() prevented the subsequent cumsum from succeeding, so I left it out:
outcome <- data_b %>%
group_by(year, treat) %>%
do(data.frame(year=.$year,
treat=.$treat,
days=seq(.$start_date, .$end_date, by="days"))) %>%
# here's the "merge" with `x$days` matched to `y$date`
# that will omit non-matching dates from data_a
left_join(data_a, by=c("days" = "date")) %>%
mutate(cum_x = cumsum(variable_x))
# decided to leave in variable _x but you could drop that col if you wanted:
# %>%select(-variable_x)
> outcome
# A tibble: 14 × 5
# Groups: year, treat [3]
year treat days variable_x cum_x
<chr> <chr> <date> <dbl> <dbl>
1 year2019 treatA 2019-01-02 14 14
2 year2019 treatA 2019-01-03 15 29
3 year2019 treatA 2019-01-04 13 42
4 year2019 treatA 2019-01-05 12 54
5 year2020 treatA 2020-01-03 11 11
6 year2020 treatA 2020-01-04 13 24
7 year2020 treatA 2020-01-05 10 34
8 year2020 treatA 2020-01-06 11 45
9 year2021 treatB 2021-01-03 14 14
10 year2021 treatB 2021-01-04 14 28
11 year2021 treatB 2021-01-05 12 40
12 year2021 treatB 2021-01-06 13 53
13 year2021 treatB 2021-01-07 13 66
14 year2021 treatB 2021-01-08 11 77