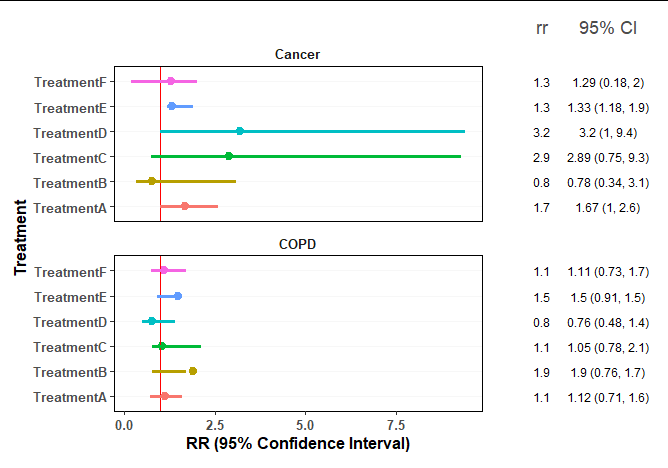
I have created a forest plot and am trying to change the colours of specific lines on my plot (just for preference). For example, I would like Treatment B to be light purple/lavender color and Treatment A to be a bright orange. Below is my code but I haven't been able to figure out where/how to adjust it to specify colors. Below is also an example of my current plot that I want to change. Any help is appreciated!
tester <- data.frame(
treatmentgroup = c("TreatmentA", "TreatmentB", "TreatmentC", "TreatmentD", "TreatmentE", "TreatmentF", "TreatmentA", "TreatmentB", "TreatmentC", "TreatmentD", "TreatmentE", "TreatmentF"),
rr = c(1.12, 1.9, 1.05, 0.76, 1.5, 1.11, 1.67, 0.78, 2.89, 3.2, 1.33, 1.29),
low_ci = c(0.71, 0.76, 0.78, 0.48, 0.91, 0.73, 1, 0.34, 0.75, 1, 1.18, 0.18),
up_ci = c(1.6, 1.7, 2.11, 1.4, 1.5, 1.7, 2.6, 3.1, 9.3, 9.4, 1.9, 2),
RR_ci = c(
"1.12 (0.71, 1.6)", "1.9 (0.76, 1.7)", "1.05 (0.78, 2.1)", "0.76 (0.48, 1.4)", "1.5 (0.91, 1.5)", "1.11 (0.73, 1.7)",
"1.67 (1, 2.6)", "0.78 (0.34, 3.1)", "2.89 (0.75, 9.3)", "3.2 (1, 9.4)", "1.33 (1.18, 1.9)", "1.29 (0.18, 2)"
),
ci = c(
"0.71, 1.6",
"0.76, 1.7",
"0.78, 2.1",
"0.48, 1.4",
"0.91, 1.5",
"0.73, 1.7",
"1, 2.6",
"0.34, 3.1",
"0.75, 9.3",
"1, 9.4",
"1.18, 1.9",
"0.18, 2"
),
X = c("COPD", "COPD", "COPD", "COPD", "COPD", "COPD", "Cancer", "Cancer", "Cancer", "Cancer", "Cancer", "Cancer"),
no = c(1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12)
)
# Reduce the opacity of the grid lines: Default is 255
col_grid <- rgb(235, 235, 235, 100, maxColorValue = 255)
library(dplyr, warn = FALSE)
library(ggplot2)
library(patchwork)
forest <- ggplot(
data = tester,
aes(x = treatmentgroup, y = rr, ymin = low_ci, ymax = up_ci)
)
geom_pointrange(aes(col = treatmentgroup))
geom_hline(yintercept = 1, colour = "red")
xlab("Treatment")
ylab("RR (95% Confidence Interval)")
geom_errorbar(aes(ymin = low_ci, ymax = up_ci, col = treatmentgroup), width = 0, cex = 1)
facet_wrap(~X, strip.position = "top", nrow = 9, scales = "free_y")
theme_classic()
theme(
panel.background = element_blank(), strip.background = element_rect(colour = NA, fill = NA),
strip.text.y = element_text(face = "bold", size = 12),
panel.grid.major.y = element_line(colour = col_grid, size = 0.5),
strip.text = element_text(face = "bold"),
panel.border = element_rect(fill = NA, color = "black"),
legend.position = "none",
axis.text = element_text(face = "bold"),
axis.title = element_text(face = "bold"),
plot.title = element_text(face = "bold", hjust = 0.5, size = 13)
)
coord_flip()
dat_table <- tester %>%
select(treatmentgroup, X, RR_ci, rr) %>%
mutate(rr = sprintf("%0.1f", round(rr, digits = 1))) %>%
tidyr::pivot_longer(c(rr, RR_ci), names_to = "stat") %>%
mutate(stat = factor(stat, levels = c("rr", "RR_ci")))
table_base <- ggplot(dat_table, aes(stat, treatmentgroup, label = value))
geom_text(size = 3)
scale_x_discrete(position = "top", labels = c("rr", "95% CI"))
facet_wrap(~X, strip.position = "top", ncol = 1, scales = "free_y", labeller = labeller(X = c(Cancer = "", COPD = "")))
labs(y = NULL, x = NULL)
theme_classic()
theme(
strip.background = element_blank(),
panel.grid.major = element_blank(),
panel.border = element_blank(),
axis.line = element_blank(),
axis.text.y = element_blank(),
axis.text.x = element_text(size = 12),
axis.ticks = element_blank(),
axis.title = element_text(face = "bold"),
)
forest table_base plot_layout(widths = c(10, 4))
CodePudding user response:
Use scale_colour_manual() and provide a named vector of colours.
E.g. scale_colour_manual(values = c("TreatmentA"= "red", "TreatmentB" = "orange")