I created a cuminc object and can print the estimates and variances directly in my console. I then used ggcompetingrisks to create a plot, and used the option conf.int = TRUE to plot the confidence intervals. However, I can't find the values of the confidence intervals.
CodePudding user response:
The confidence bounds are not saved in the output. Fortunately it's easy to calculate them manually. Just extract est and std from plot$data and calculate est 1.96*std and est-1.96*std, replacing 1.96 with other z-values for other confidence limits.
library(survminer)
library(cmprsk)
set.seed(2)
ss <- rexp(100)
gg <- factor(sample(1:3,100,replace=TRUE),1:3,c('BRCA','LUNG','OV'))
cc <- factor(sample(0:2,100,replace=TRUE),0:2,c('no event', 'death', 'progression'))
strt <- sample(1:2,100,replace=TRUE)
fit <- cmprsk::cuminc(ss,cc,gg,strt)
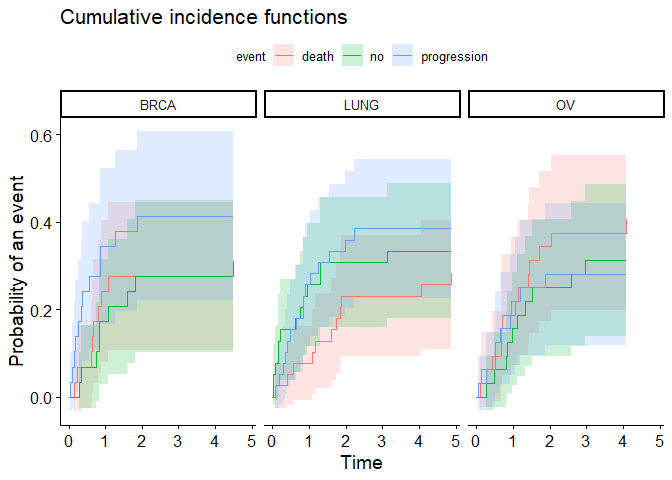
plot <- ggcompetingrisks(fit, conf.int = TRUE)
plot
# Calculate confidence bounds manually
dat <- plot$data
dat$ymax <- dat$est 1.96*dat$std
dat$ymin <- dat$est - 1.96*dat$std
head(dat)
#> time est var name event group std
#> 1 0.0000000 0.00000000 0.000000000 BRCA no event no BRCA 0.00000000
#> 2 0.2731330 0.00000000 0.000000000 BRCA no event no BRCA 0.00000000
#> 3 0.2731330 0.03448276 0.001202639 BRCA no event no BRCA 0.03467908
#> 4 0.3312168 0.03448276 0.001202639 BRCA no event no BRCA 0.03467908
#> 5 0.3312168 0.06896552 0.002321886 BRCA no event no BRCA 0.04818595
#> 6 0.7388502 0.06896552 0.002321886 BRCA no event no BRCA 0.04818595
#> ymax ymin
#> 1 0.0000000 0.00000000
#> 2 0.0000000 0.00000000
#> 3 0.1024538 -0.03348824
#> 4 0.1024538 -0.03348824
#> 5 0.1634100 -0.02547895
#> 6 0.1634100 -0.02547895
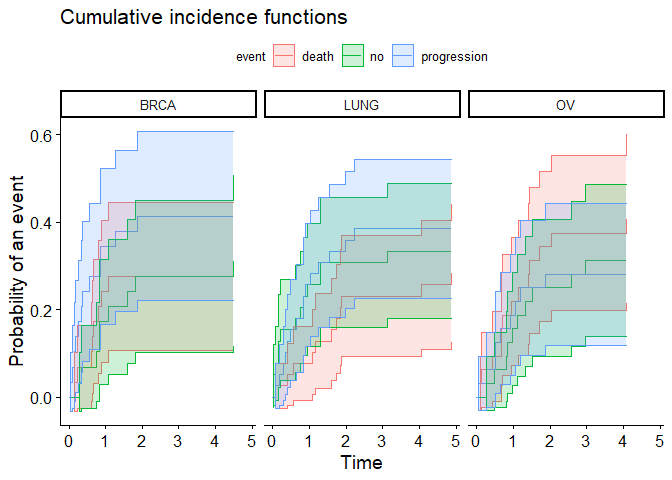
# Plot to confirm they match
ggcompetingrisks(fit, conf.int = FALSE)
geom_ribbon(data = dat,
mapping = aes(ymin = ymin, ymax = ymax, x = time,
fill = event),
alpha = 0.2)
Created on 2022-09-13 with reprex v2.0.2
