With the R code below,
library(ggplot2)
library(ggridges)
ggplot(iris, aes(x = Sepal.Length, y = Species))
stat_density_ridges(quantile_lines = TRUE, quantiles = c(0.025, 0.975), alpha = 0.7)
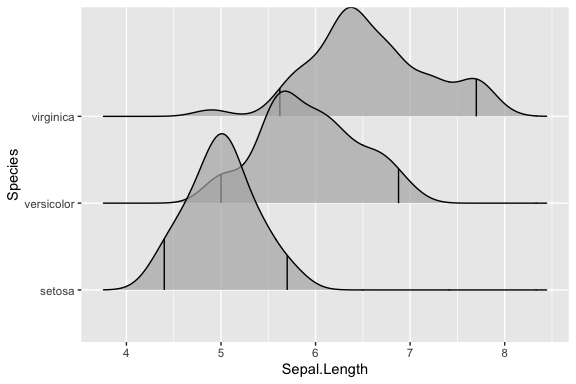
I get the following ridge plot
I would like to replace the two quantile lines (2.5% and 97.5%) under each density plot with two different lines based on the following code:
require(coda)
aggregate(Sepal.Length ~ Species, iris, function(x) HPDinterval(as.mcmc(x), 0.975))
In other words, I wish to put those vertical lines at the following locations:
Species Sepal.Length.1 Sepal.Length.2
1 setosa 4.3 5.8
2 versicolor 4.9 7.0
3 virginica 4.9 7.9
How to achieve this?
CodePudding user response:
The inverse of quantile is ecdf, so you can back-transform your lengths into quantiles to feed to stat_density_ridges:
aggregate(Sepal.Length ~ Species, iris, function(x) {
ecdf(x)(HPDinterval(as.mcmc(x), 0.975))})
#> Species Sepal.Length.1 Sepal.Length.2
#> 1 setosa 0.02 1.00
#> 2 versicolor 0.02 1.00
#> 3 virginica 0.02 1.00
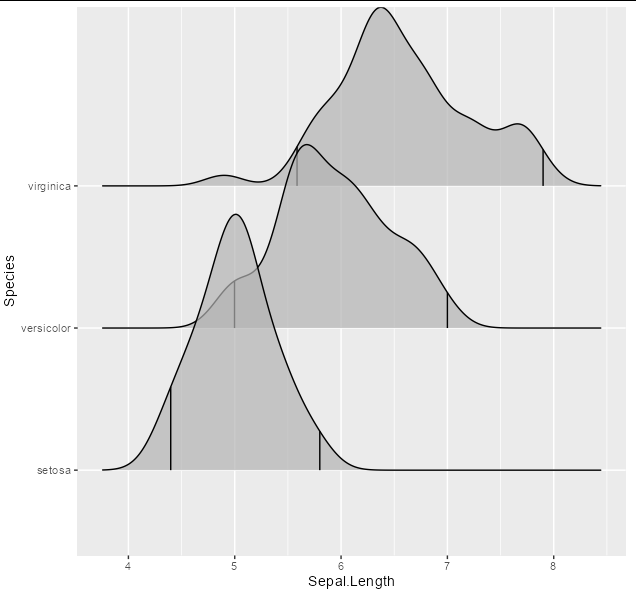
And you will see that the quantiles you are looking for are 0.02 and 1, so now you can plot:
ggplot(iris, aes(x = Sepal.Length, y = Species))
stat_density_ridges(quantile_lines = TRUE, quantiles = c(0.02, 1),
alpha = 0.7)
Note also that the function you are passing to aggregate is the same as passing range
aggregate(Sepal.Length ~ Species, iris, range)
#> Species Sepal.Length.1 Sepal.Length.2
#> 1 setosa 4.3 5.8
#> 2 versicolor 4.9 7.0
#> 3 virginica 4.9 7.9
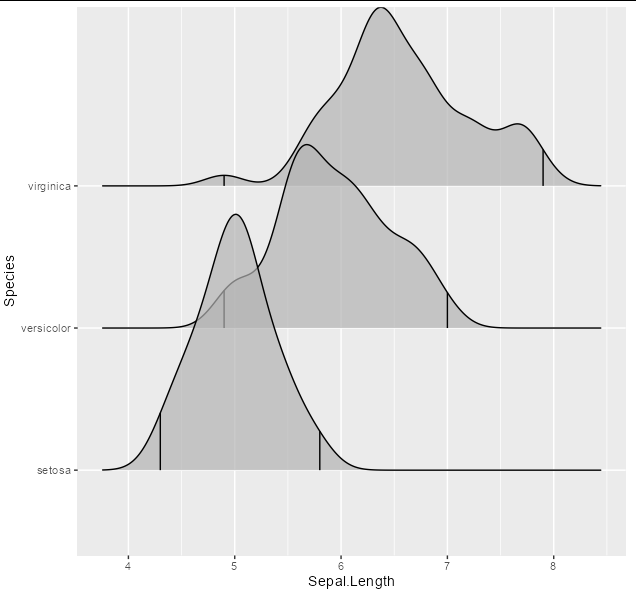
So you could simply pass 0 and 1 as quantiles, which matches the numbers in your table perfectly:
ggplot(iris, aes(x = Sepal.Length, y = Species))
stat_density_ridges(quantile_lines = TRUE, quantiles = c(0, 1),
alpha = 0.7)
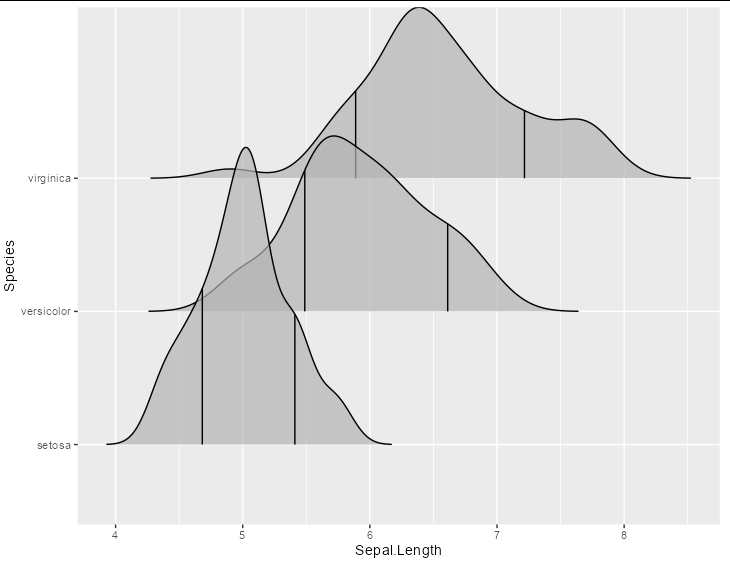
Edit
If the quantiles returned by your function are all different, you could use multiple geom layers, which you could have in a list, like this:
library(ggridges)
ggplot(iris, aes(x = Sepal.Length, y = Species))
lapply(split(iris, iris$Species)[3:1], function(x) {
stat_density_ridges(quantile_lines = TRUE,
quantiles = ecdf(x$Sepal.Length)(HPDinterval(as.mcmc(x$Sepal.Length), 0.75)),
alpha = 0.7, data = x)})