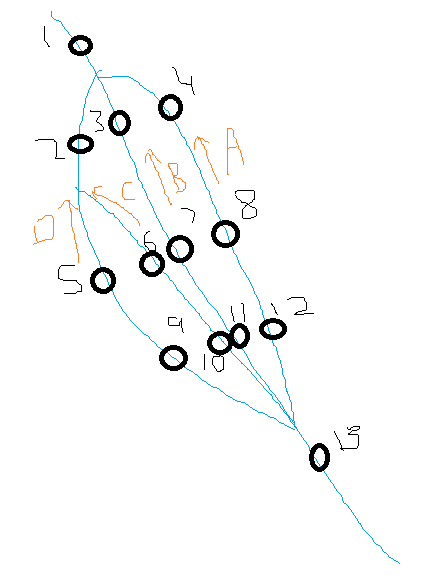
I am trying to come up with a way to loop through a data frame and recognise points shared between 2 columns and work iteratively to assign a unique factor to these. Specifically, I have a data frame indicating points along a river and which points are immediately upstream of this.
Here is some example data:
df <- data.frame(RiverID = rep(c(1,2), each = 15), SiteID = rep((c(2,3,4,5,6,7,8,9,10,11,12,13,13,13,13)),2),
Upstream_SiteID = rep((c(1,1,1,2,2,3,4,5,6,7,8,9,10,11,12)),2),
Dist2Mouth = rep((c(2000,2000,2000,1500,1500,1500,1500,1000,1000,1000,1000,500,500,500,500)),2))
And the basic paint image below might help explain the kind of data I have.
What I would like to do is identify all possible 'routes' through the system (orange letters in the image). So in the example, I would start from point 13, go 'upstream' in the dataframe (i.e. lower Distance2Mouth values) and recognise 4 different routes (A-D). I then need to iteratively keep working up the data frame and assign routes to each point.
There are some instances where the stretch of river between the two points could belong to two routes. For example in the image below, the section between 2-1 could be part of routes C or D. In these instances, I would like to create multiple rows with the same SiteID and Upstream_SiteID that list the different potential routes.
Lastly, I have these instances across various rivers, so I would like to loop through the dataframe and apply the above code for each RiverID.
Desired output (the correct routes apply to RiverID '1'):
output <- data.frame(RiverID = rep(c(1,2), each = 16),
SiteID = rep((c(2,2,3,4,5,6,7,8,9,10,11,12,13,13,13,13)),2),
Upstream_SiteID = rep((c(1,1,1,1,2,2,3,4,5,6,7,8,9,10,11,12)),2),
Dist2Mouth = rep((c(2000,2000,2000,2000,1500,1500,1500,1500,1000,1000,1000,1000,500,500,500,500)),2),
Route = as.factor(c(rep(c("D","C","B","A"),times = 4),rep(c("H","G","F","E"),times = 4))))
CodePudding user response:
igraph is definitely the right tool for problems like this. However, it is also possible to do what you need with a few lines of code without the package. The routes are identified by the go_up() function which uses a depth-first search algorithm.
## Builds a nested list with all possible paths on its deepest level
#
# path path so far
# ups list of upstream points for each point
#
go_up <- function(path, ups) {
# last point of the path
last <- tail(path, 1)
if (last %in% names(ups)) {
# continue with all possible upstream points
lapply(ups[[as.character(last)]],
function(up) go_up(c(path, up), ups))
# finish if no upstream point exists
} else paste(path, collapse='---')
# path is collapsed into a string so that the resulting list can be
# easily flattened
}
This can be then applied to each river separately like this:
river.routes <- lapply(split(df, ~RiverID), function(river) {
# list with upstream points for each point
ups <- tapply(river$Upstream_SiteID, river$SiteID, c)
# we will start from the highest ID
last <- max(river$SiteID)
# find the routes
routes <- go_up(last, ups)
# flatten the list and split the routes into points
routes <- strsplit(unlist(routes), '---')
# add a logical column for each route
for (i in seq_along(routes)) {
river[[paste0('route', i)]] <- river$SiteID %in% routes[[i]]
}
river
})
Output:
river.routes
# $`1`
# RiverID SiteID Upstream_SiteID Dist2Mouth route1 route2 route3 route4
# 1 1 2 1 2000 TRUE TRUE FALSE FALSE
# 2 1 3 1 2000 FALSE FALSE TRUE FALSE
# 3 1 4 1 2000 FALSE FALSE FALSE TRUE
# 4 1 5 2 1500 TRUE FALSE FALSE FALSE
# 5 1 6 2 1500 FALSE TRUE FALSE FALSE
# 6 1 7 3 1500 FALSE FALSE TRUE FALSE
# 7 1 8 4 1500 FALSE FALSE FALSE TRUE
# 8 1 9 5 1000 TRUE FALSE FALSE FALSE
# 9 1 10 6 1000 FALSE TRUE FALSE FALSE
# 10 1 11 7 1000 FALSE FALSE TRUE FALSE
# 11 1 12 8 1000 FALSE FALSE FALSE TRUE
# 12 1 13 9 500 TRUE TRUE TRUE TRUE
# 13 1 13 10 500 TRUE TRUE TRUE TRUE
# 14 1 13 11 500 TRUE TRUE TRUE TRUE
# 15 1 13 12 500 TRUE TRUE TRUE TRUE
#
# $`2`
# RiverID SiteID Upstream_SiteID Dist2Mouth route1 route2 route3 route4
# 16 2 2 1 2000 TRUE TRUE FALSE FALSE
# 17 2 3 1 2000 FALSE FALSE TRUE FALSE
# 18 2 4 1 2000 FALSE FALSE FALSE TRUE
# 19 2 5 2 1500 TRUE FALSE FALSE FALSE
# 20 2 6 2 1500 FALSE TRUE FALSE FALSE
# 21 2 7 3 1500 FALSE FALSE TRUE FALSE
# 22 2 8 4 1500 FALSE FALSE FALSE TRUE
# 23 2 9 5 1000 TRUE FALSE FALSE FALSE
# 24 2 10 6 1000 FALSE TRUE FALSE FALSE
# 25 2 11 7 1000 FALSE FALSE TRUE FALSE
# 26 2 12 8 1000 FALSE FALSE FALSE TRUE
# 27 2 13 9 500 TRUE TRUE TRUE TRUE
# 28 2 13 10 500 TRUE TRUE TRUE TRUE
# 29 2 13 11 500 TRUE TRUE TRUE TRUE
# 30 2 13 12 500 TRUE TRUE TRUE TRUE
I didn't much like the idea of duplicating the rows for points located on more than one route so I rather added a logical column for each route instead. You can change that if you like, the routes are available inside the lapply in the routes variable which looks like this:
# [[1]]
# [1] "13" "9" "5" "2" "1"
#
# [[2]]
# [1] "13" "10" "6" "2" "1"
#
# [[3]]
# [1] "13" "11" "7" "3" "1"
#
# [[4]]
# [1] "13" "12" "8" "4" "1"
CodePudding user response:
The core function you want with igraph is all_simple_paths which will give you a list of paths/routes. To create a graph, you need unique ids, so I'm just ignoring the second river piece.
library(igraph)
library(purrr)
library(dplyr)
df_w1 <- data.frame(
SiteID = c(2,3,4,5,6,7,8,9,10,11,12,13,13,13,13),
Upstream_SiteID = c(1,1,1,2,2,3,4,5,6,7,8,9,10,11,12),
Dist2Mouth = c(2000,2000,2000,1500,1500,1500,1500,1000,1000,1000,1000,500,500,500,500)
)
paths <- graph_from_data_frame(df_w1) |> # create graph object
all_simple_paths("13", "1") |> # enumerate paths
map(~attr(.x, "names") |> as.integer()) |> # get the labels and not internal ids
print()
#> [[1]]
#> [1] 13 9 5 2 1
#>
#> [[2]]
#> [1] 13 10 6 2 1
#>
#> [[3]]
#> [1] 13 11 7 3 1
#>
#> [[4]]
#> [1] 13 12 8 4 1
For your purposes then you can enumerate the routes each segment belongs to and then join back to the original data (as not to lose the metadata). Iterate through the segments and check to see if both sites are in each of the paths.
# assume less than 26 routes
codingList <- map2_dfr(
df_w1$SiteID, df_w1$Upstream_SiteID,
\(from, to) tibble(
SiteID = from,
Upstream_SiteID = to,
Route = LETTERS[map_lgl(paths, \(p) all(c(from, to) %in% p)) |> which()]
)
) |>
print()
#> # A tibble: 16 × 3
#> SiteID Upstream_SiteID Route
#> <dbl> <dbl> <chr>
#> 1 2 1 A
#> 2 2 1 B
#> 3 3 1 C
#> 4 4 1 D
#> 5 5 2 A
#> 6 6 2 B
#> 7 7 3 C
#> 8 8 4 D
#> 9 9 5 A
#> 10 10 6 B
#> 11 11 7 C
#> 12 12 8 D
#> 13 13 9 A
#> 14 13 10 B
#> 15 13 11 C
#> 16 13 12 D
df_w1 |>
left_join(codingList, by = c("SiteID", "Upstream_SiteID"))
#> SiteID Upstream_SiteID Dist2Mouth Route
#> 1 2 1 2000 A
#> 2 2 1 2000 B
#> 3 3 1 2000 C
#> 4 4 1 2000 D
#> 5 5 2 1500 A
#> 6 6 2 1500 B
#> 7 7 3 1500 C
#> 8 8 4 1500 D
#> 9 9 5 1000 A
#> 10 10 6 1000 B
#> 11 11 7 1000 C
#> 12 12 8 1000 D
#> 13 13 9 500 A
#> 14 13 10 500 B
#> 15 13 11 500 C
#> 16 13 12 500 D