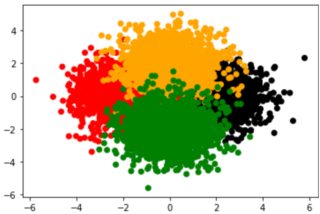
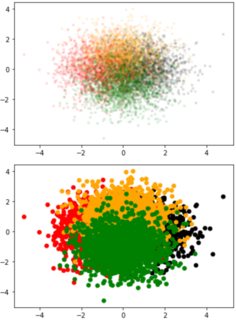
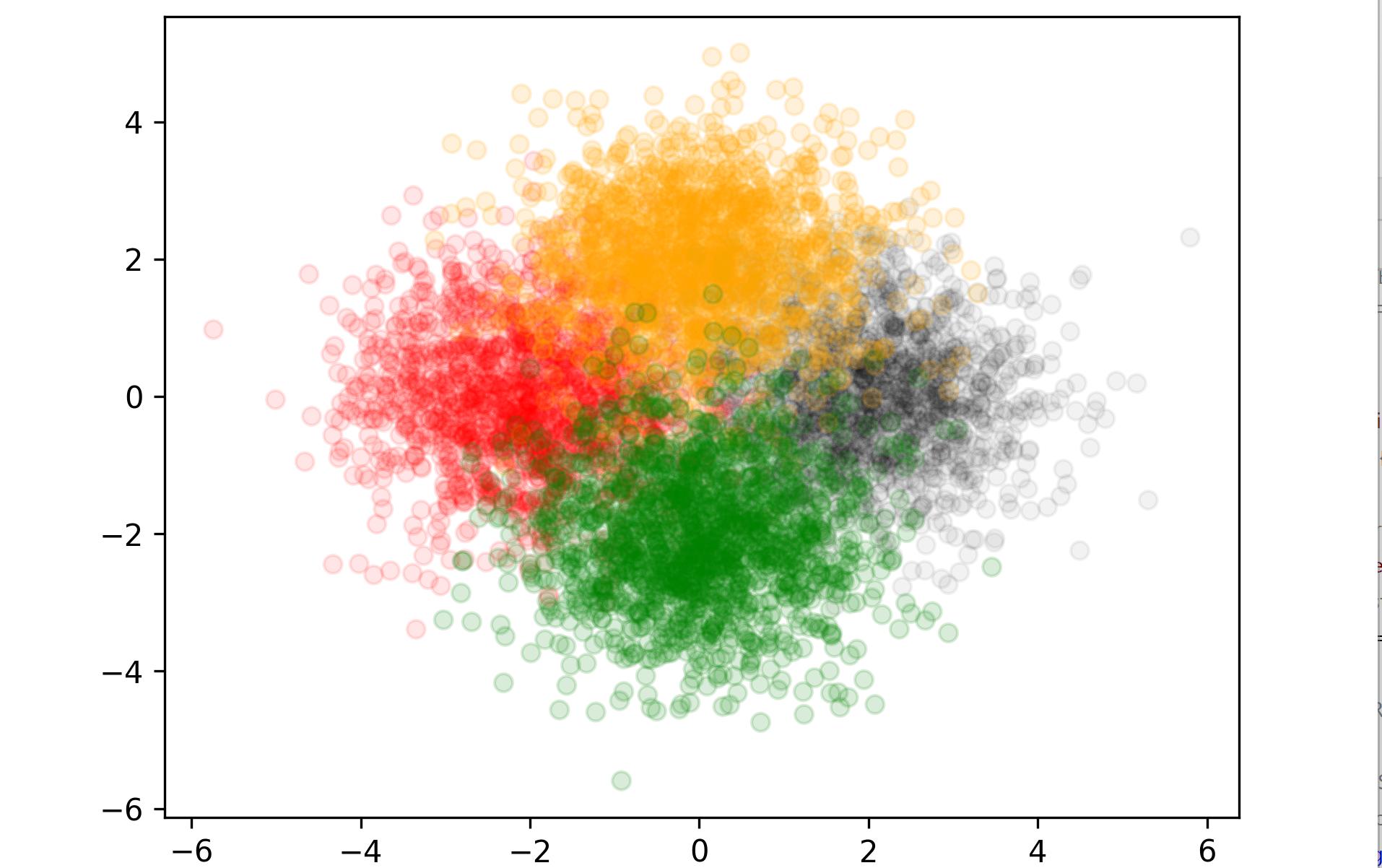
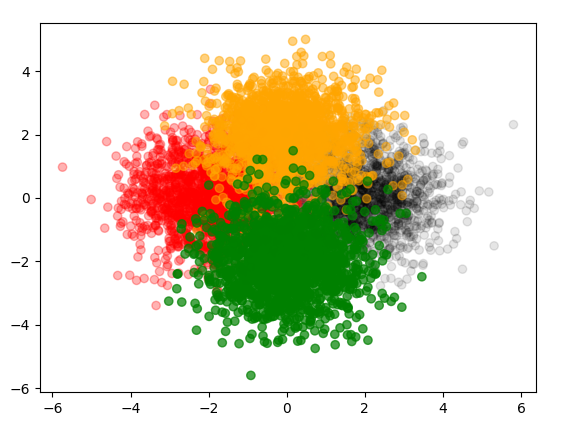
I have the following scatter plot and I want to decrease the transparency of each color to see all the clusters clearly:
import numpy as np
import matplotlib.pyplot as plt
n = 1600
np.random.seed(0)
data1 = np.random.randn(n, 2) np.asarray([2, 0])
data2 = np.random.randn(n, 2) np.asarray([-2, 0])
data3 = np.random.randn(n, 2) np.asarray([0, 2])
data4 = np.random.randn(n, 2) np.asarray([0, -2])
data = np.concatenate((data1, data2, data3, data4))
labels = np.concatenate((n*[1], n*[2], n*[3], n*[4]))
fig, ax = plt.subplots()
colors_list = ['k', 'r', 'orange', 'g']
color_codes = np.asarray(colors_list)
colors = color_codes[labels-1]
ax.scatter(*data.T, c = colors);
Question
How can I do it without For loop in an efficient way?
Edit
n = 1600
np.random.seed(0)
data1 = np.random.randn(n, 2) np.asarray([1, 0])
data2 = np.random.randn(n, 2) np.asarray([-1, 0])
data3 = np.random.randn(n, 2) np.asarray([0, 1])
data4 = np.random.randn(n, 2) np.asarray([0, -1])
data = np.concatenate((data1, data2, data3, data4))
labels = np.concatenate((n*[1], n*[2], n*[3], n*[4]))
# alphas = np.concatenate((n*[.1], n*[.2], n*[.3], n*[.4]))
alphas = np.linspace(0.1, 1, n)
fig, ax = plt.subplots()
colors_list = ['k', 'r', 'orange', 'g']
color_codes = np.asarray(colors_list)
colors = color_codes[labels-1]
ax.scatter(*data.T, c = colors, alpha = 0.1, s=10);
fig, ax = plt.subplots()
colors_list = ['k', 'r', 'orange', 'g']
color_codes = np.asarray(colors_list)
colors = color_codes[labels-1]
ax.scatter(*data.T, c = colors);
Problem
The black cluster is totally overlaid and I cannot see it in the second picture. Is there a way that I can use and find a clear picture that helps me to see all the colors?
CodePudding user response:
To set the same transparency to all, you can use ax.scatter(*data.T, c=colors, alpha=0.1).
To have individual transparencies, you could work with to_rgba to include transparency into the colors.
import numpy as np
import matplotlib.pyplot as plt
from matplotlib.colors import to_rgba
n = 1600
np.random.seed(0)
data1 = np.random.randn(n, 2) np.asarray([2, 0])
data2 = np.random.randn(n, 2) np.asarray([-2, 0])
data3 = np.random.randn(n, 2) np.asarray([0, 2])
data4 = np.random.randn(n, 2) np.asarray([0, -2])
data = np.concatenate((data1, data2, data3, data4))
labels = np.concatenate((n * [1], n * [2], n * [3], n * [4]))
labels = np.repeat([1, 2, 3, 4], n)
fig, ax = plt.subplots()
colors_list = [to_rgba('k', 0.05), to_rgba('r', 0.1), to_rgba('orange', 0.15), to_rgba('g', 0.15)]
color_codes = np.asarray(colors_list)
colors = color_codes[labels - 1]
ax.scatter(*data.T, c=colors)
plt.show()
CodePudding user response:
Your code is almost there, and @JohanC gave an answer that can be followed up upon in the same way you apply your colors. Create a list of alphas and pass them to plt.scatter
import numpy as np
import matplotlib.pyplot as plt
n = 1600
np.random.seed(0)
data1 = np.random.randn(n, 2) np.asarray([2, 0])
data2 = np.random.randn(n, 2) np.asarray([-2, 0])
data3 = np.random.randn(n, 2) np.asarray([0, 2])
data4 = np.random.randn(n, 2) np.asarray([0, -2])
data = np.concatenate((data1, data2, data3, data4))
labels = np.concatenate((n*[1], n*[2], n*[3], n*[4]))
fig, ax = plt.subplots()
colors_list = ['k', 'r', 'orange', 'g']
alpha_list = [0.1,0.3,0.5,0.7]
color_codes = np.asarray(colors_list)
colors = color_codes[labels-1]
alphas = np.asarray(alpha_list)[labels-1]
ax.scatter(*data.T, c = colors, alpha = alphas);
plt.show()
Just be sure that the alpha array is the same length as the color array.
For additional clarity, it may be worth reducing the size of the scatter points with the s=<size> keyword in scatter. The edges in each marker can also be disabled. Lastly, for scatter plots that have highly localized density, it is usually better to plot the points as a 2D histogram or hexbinned histogram. Hope this helped!