How does one change the size of the points only in a ggplot2 legend? I want them to remain the size I have in the plot code, but to appear bigger in the legend.
My plot code is here:
ggplot(df, aes(x=log(length), y=log(wt), shape=Depth,fill=Depth,color=Depth, linetype=Depth))
geom_point(aes(),size=2,alpha=1,stroke=1.3)
geom_smooth(method=lm, se=FALSE, fullrange=FALSE)
facet_wrap(~ LakeYear, nrow=3, ncol=2)
labs(x="ln Length (mm)", y="ln Weight (mg)")
scale_fill_manual(values=cbPalette2,name="Depth")
scale_color_manual(values=cbPalette,name="Depth")
scale_shape_manual(values=c(16,15,21,18),name="Depth")
scale_linetype_manual(values =c("<5 m"="solid", "10-15 m"="solid", "15-20 m"="solid",">20 m"="dashed"),name="Depth")
PlotTheme
theme(panel.spacing.x=unit(1.4, "lines"),panel.spacing.y=unit(1, "lines"),legend.title=element_text(size=16))
I have tried to add guides(color = guide_legend(override.aes = list(size=3))) and guides(shape = guide_legend(override.aes = list(size = 2))) but this just changes the line size. I have tried some code I found from an example 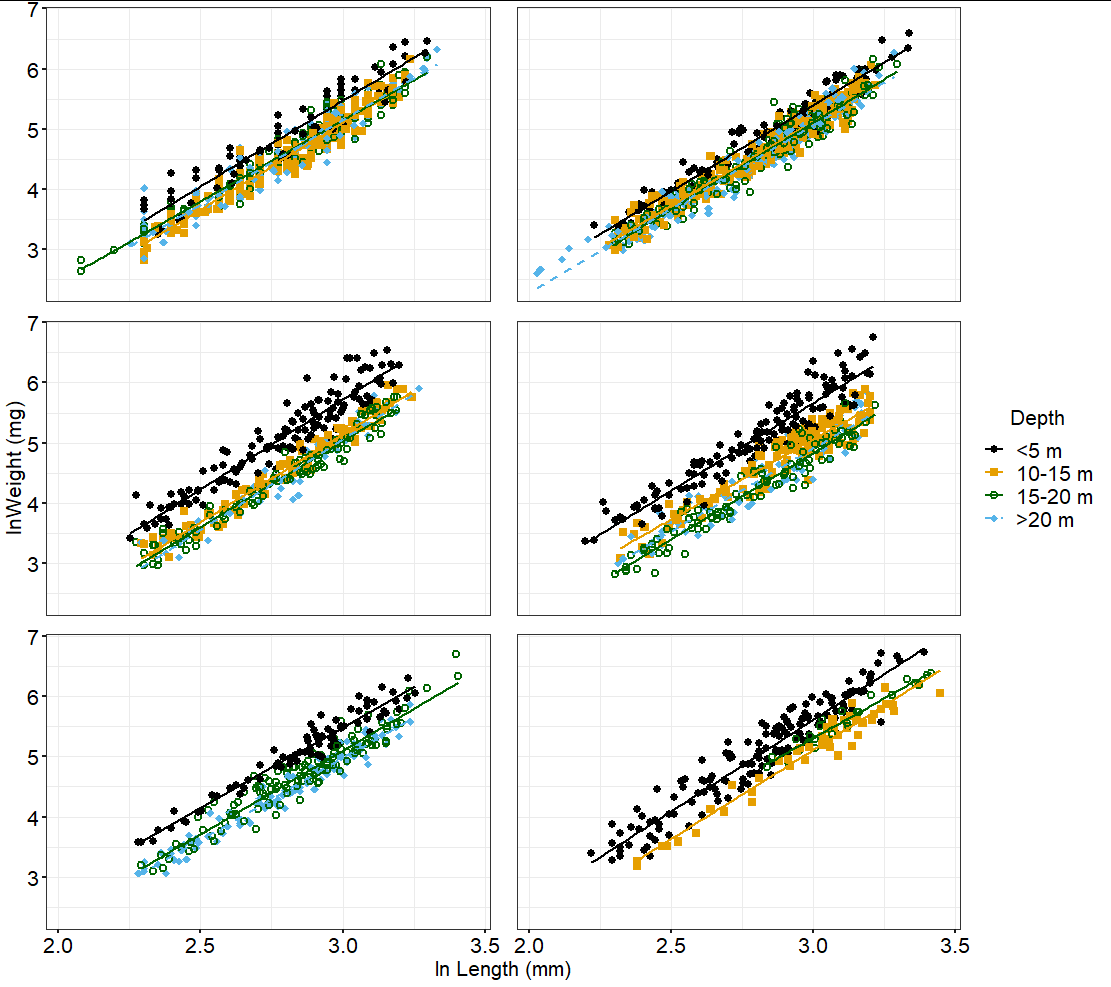
Added either
p guides(color = guide_legend(override.aes = list(size=5)))
p guides(shape = guide_legend(override.aes = list(size = 5)))
I get this, without the actual shape changed, only the line
CodePudding user response:
The simplest answer seems to be making two versions of the same plot. The first is the initial plot you want with the point sizes you'd like in the main plot. The second plot is used to grab only the legend, so you increase the point sizes to where you want them to be in the legend. You then use cowplot::get_legend and cowplot::plot_grid to extract the legend and arrange the plots together as you'd like them.
FinalFigure<-cowplot::plot_grid(
Figure_Initial theme(legend.position = "none"),
cowplot::get_legend(Figure_LegendGrab),
ncol = 2, rel_widths = c(4, 0.8))
CodePudding user response:
Vanilla ggplot can get really really hacky when you have very specific ideas to modify your legend.
In those cases, it is often way easier to create a fake legend and stick it to your main plot. It's not fully automatic, but largely so, and requires (minor) fiddling with some spacing. But I think this should work for most of the graphs where such special modifications are needed - they will hopefully not be many. Having said that, if you need this type of visualisation on a large scale, maybe you want to reconsider your visualisation.
library(tidyverse)
library(patchwork)
## df <- ... your data df
## your plot, simplified
p <- ggplot(df, aes(x = log(length), y = log(wt), shape = Depth, color = Depth))
geom_point()
geom_smooth(method = lm, se = FALSE)
theme(legend.position = "none")
## first get the only present levels of your data
df$Depth <- droplevels(df$Depth)
# this is just a hack to center your legend more or less compared with the main plot
# change the y in order to change the distance between the legend items
ylims <- range(log(df$wt))
ymid <- quantile(ylims, c(.4,.5))
y_pos <- seq(ymid[1], ymid[2], length = length(levels(df$Depth)))
# this is just a random parameter to define the segment length
x_len <- 1.5
df_leg <- data.frame(x = 1, y = y_pos, Depth = levels(df$Depth))
p_leg <-
ggplot(df_leg, aes(x, y, color = Depth, shape = Depth))
geom_point(size = 4)
geom_segment(aes(x = x_len-x, xend = x_len, yend = y))
## bit of a
geom_text(aes(x = x_len x, label = Depth), hjust = 0.1)
## need to clip off , use your preset y limits for correct centering,
coord_cartesian(ylim= ylims, clip = "off")
theme_void()
theme(legend.position = "none",
## add a margin, otherwise you won't see your legend
plot.margin = margin(r = .5, l = .1, unit = "in"))
p p_leg
plot_layout(widths = c(1, .1))
#> `geom_smooth()` using formula 'y ~ x'
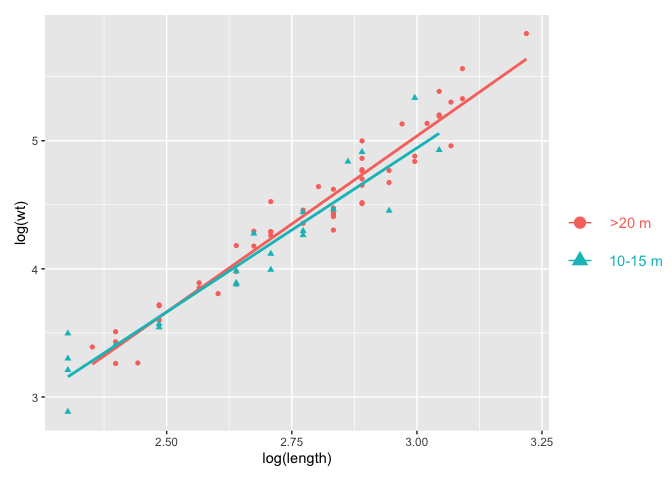
Created on 2022-10-19 by the reprex package (v2.0.1)
