## KAPLAN MEIER
fit.obj<-survfit(Surv(duree_suivi,k_oesogastr) ~ baria_t, data = pts_raw_matched)
library(survminer)
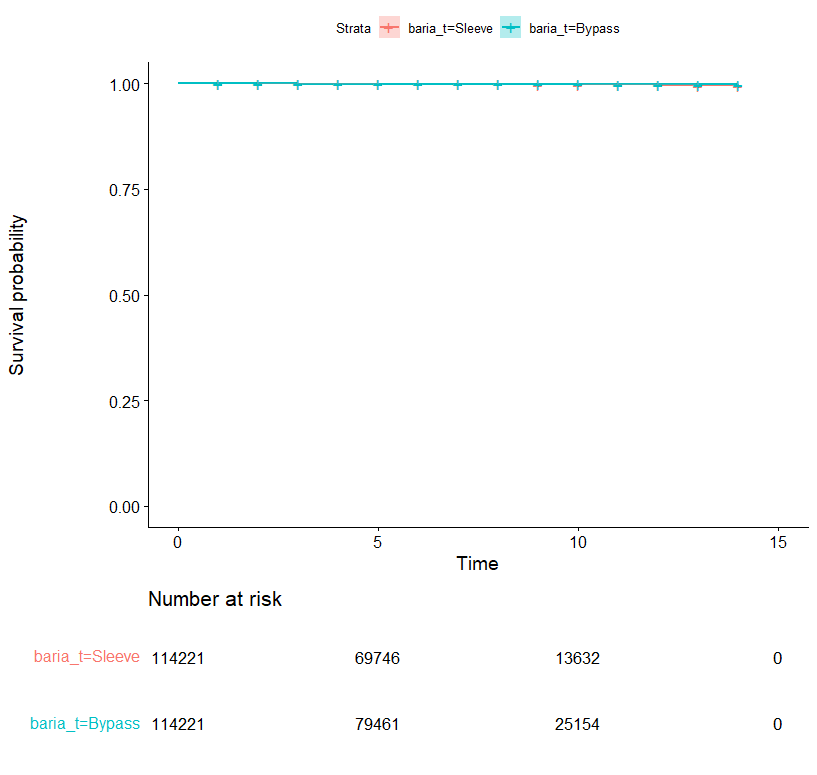
I am comparing two curves. But the number of events is so low that you can't see the curves very well. I want to truncate the y-axis (between 0.9 and 1 for example) to see the evolution of the two curves.
n events median 0.95LCL 0.95UCL
baria_t=Sleeve 114221 61 NA NA NA
baria_t=Bypass 114221 58 NA NA NA
ggsurvplot(fit.obj,
conf.int = T,
risk.table ="absolute",
tables.theme = theme_cleantable())
Here are the survival curves:
CodePudding user response:
If we use some simulated data we can replicate your problem:
set.seed(1)
pts_raw_matched <- data.frame(
duree_suivi = c(runif(114221, 0, 25000),runif(114221, 0, 26000)),
baria_t = rep(c('Sleeve', 'Bypass'), each = 114221)
)
pts_raw_matched$k_oesogastr <- as.numeric(pts_raw_matched$duree_suivi < 15)
pts_raw_matched$duree_suivi[pts_raw_matched$duree_suivi > 15] <- 15
Now we should have a data set with the same names and approximately the same characteristics as your own data.
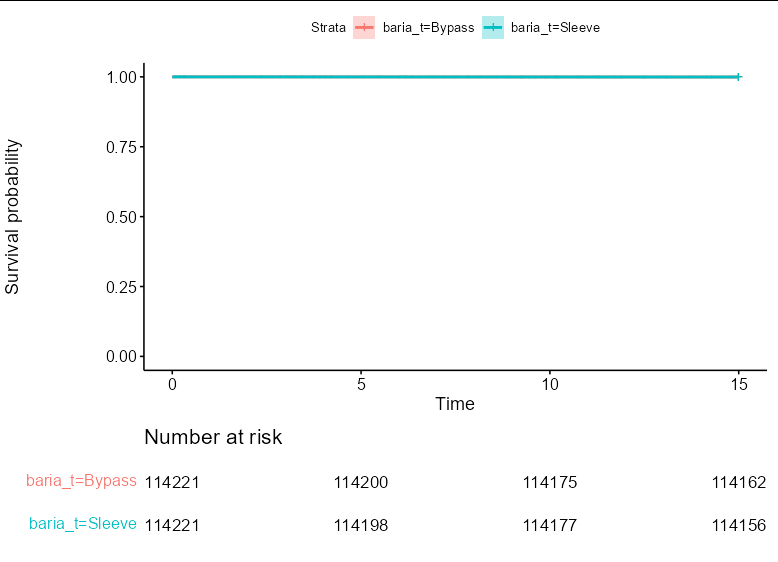
We start by creating the plot using your own code, but storing the result:
fit.obj <- survfit(Surv(duree_suivi,k_oesogastr) ~ baria_t,
data = pts_raw_matched)
ggs <- ggsurvplot(fit.obj,
conf.int = T,
risk.table ="absolute",
tables.theme = theme_cleantable())
ggs
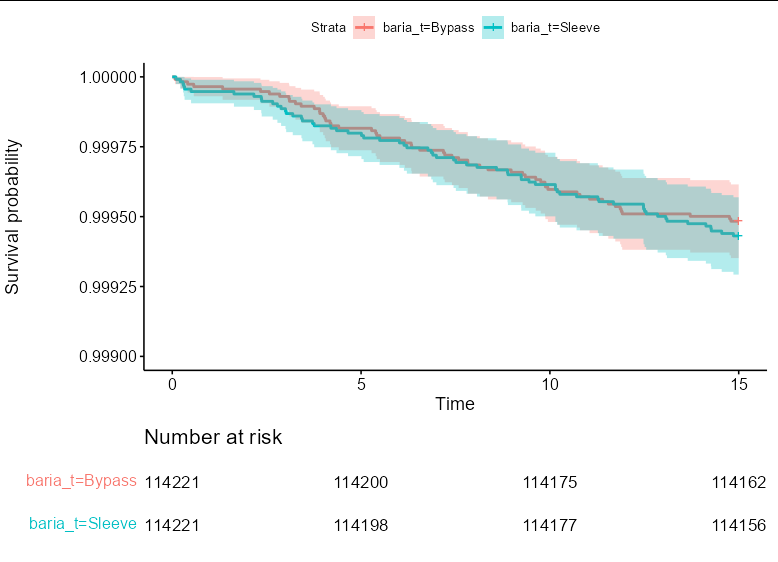
In order to change the y axis, we need to set the ylim of the plot component of the ggs object. In this case, we need to set the limits between 0.999 and 1 to be able to properly compare the curves:
ggs$plot <- ggs$plot ylim(c(0.999, 1))
ggs
CodePudding user response:
you can add argument ylim=c(0.999, 1) directly in the call to ggsurvplot:
ggsurvplot(fit.obj,
conf.int = T,
risk.table ="absolute",
tables.theme = theme_cleantable(),
ylim=c(0.999, 1))
You can do the same for xlim