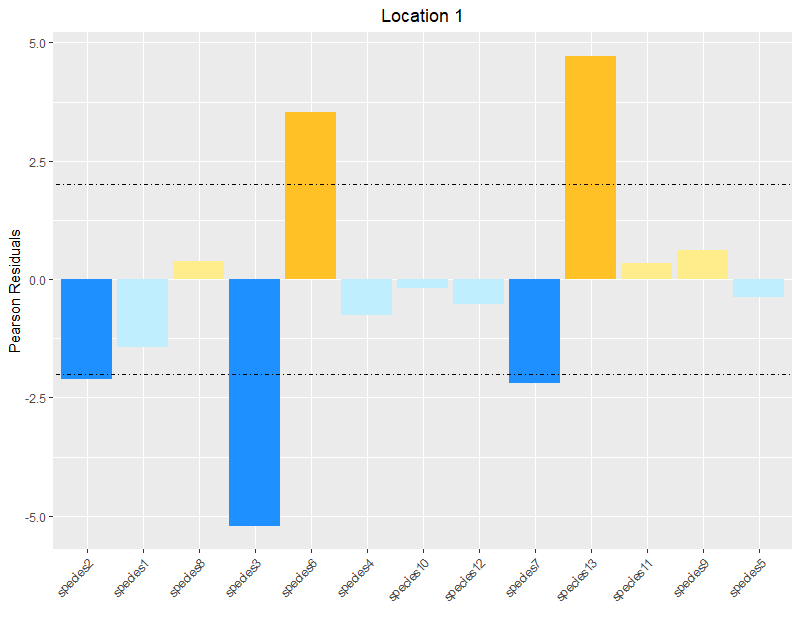
I am trying to recolor bars on a bar graph based on certain conditions of the values. (are they positive or negative? are they above or below the threshold?). Because I have do to a lot of these plots, I thought the easiest way to do that would be to create a column with the colors I want the bars to be, based on those conditions. This was easy enough with a few ifelse statements. But now, the problem is that ggplot won't pull those colors in the correct order. I have tried several different ways of doing this and can't seem to get it right.
Here is an mock-up of dataframe filtered for just the first location we want to graph, with some example data. I have provided the full dput at the bottom so you can reproduce the full example yourself.
species location test_residuals species_order color
1 species2 location1 -2.1121481 1 dodgerblue1
2 species1 location1 -1.4315793 2 lightblue1
3 species8 location1 0.3727298 3 lightgoldenrod1
4 species3 location1 -5.2163387 4 dodgerblue1
5 species6 location1 3.5301076 5 goldenrod1
6 species4 location1 -0.7546595 6 lightblue1
7 species10 location1 -0.1857843 7 lightblue1
8 species12 location1 -0.5199749 8 lightblue1
9 species7 location1 -2.1884659 9 dodgerblue1
10 species13 location1 4.7223194 10 goldenrod1
11 species11 location1 0.3374291 11 lightgoldenrod1
12 species9 location1 0.6245307 12 lightgoldenrod1
13 species5 location1 -0.3676778 13 lightblue1
when I try this
test.plot.1<- data1 %>%
filter(location == "location1") %>%
ggplot(aes(
reorder(x = species, species_order),
y= test_residuals,
fill = species))
geom_bar( stat= "identity")
ggtitle("Location 1")
theme_pubclean(
base_size = 14 )
theme(plot.title = element_text(hjust = 0.5),
legend.position = "none")
xlab("") ylab("Pearson Residuals")
scale_x_discrete(guide = guide_axis(angle = 45))
geom_abline(intercept = 2, slope = 0, linetype = "dotdash")
geom_abline(intercept = -2, slope = 0, linetype = "dotdash")
scale_fill_manual(values = color)
I get the error " Error in is_missing(values) : object 'color' not found"
If I instead specify the dataframe with:
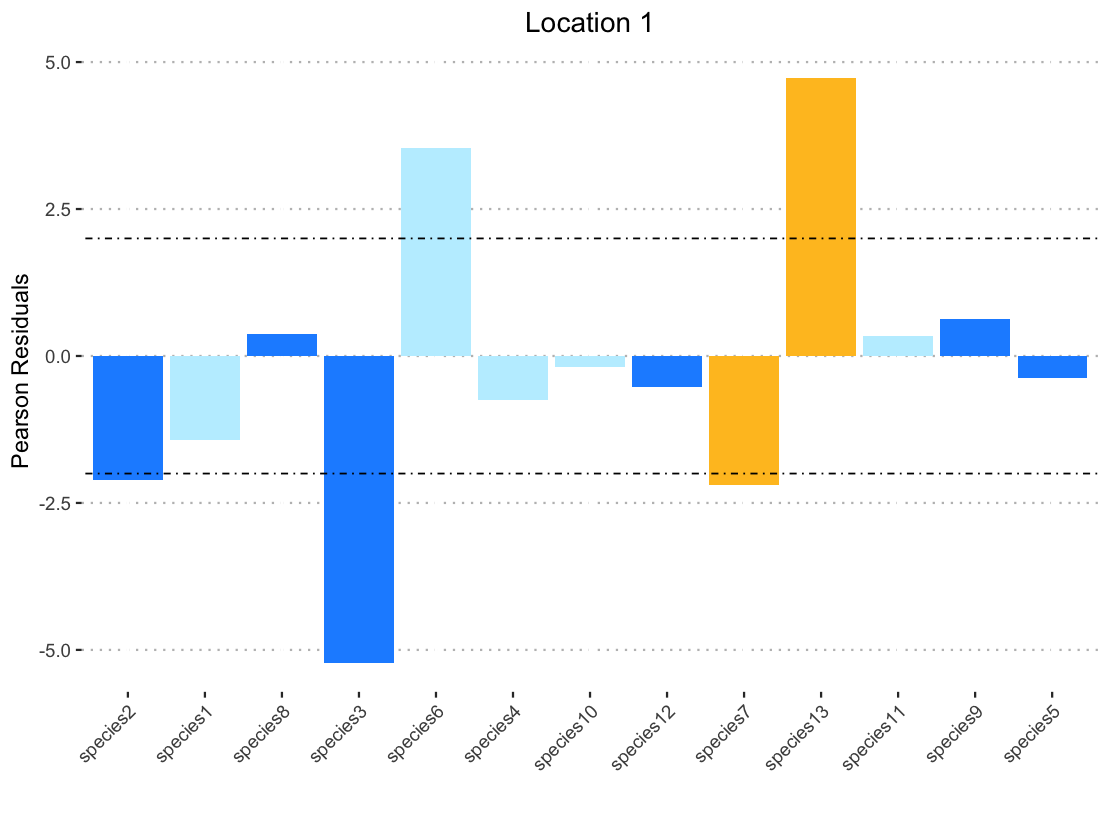
scale_fill_manual(values = data1$color)
I don't get an error, and the color pallet is even correct, but the bars themselves are not the correct color!
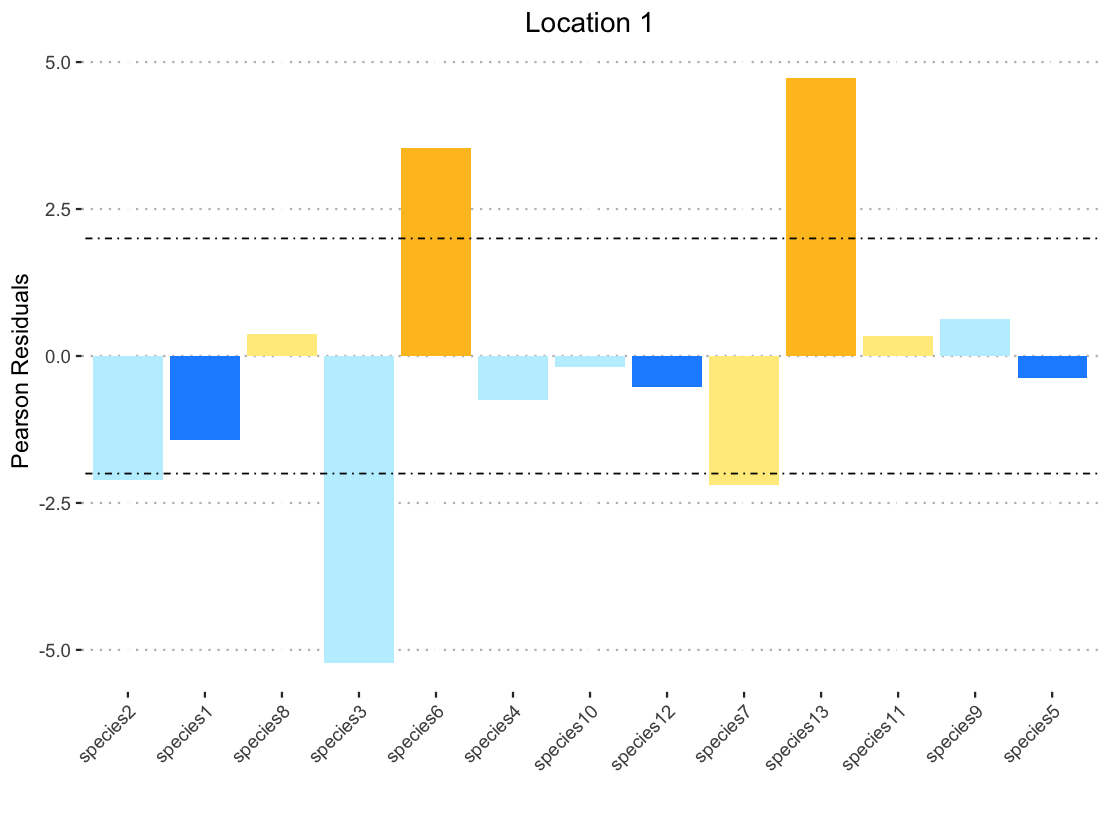
I also get miscolored bars if I specify another vector in fill (for example color) produces this:
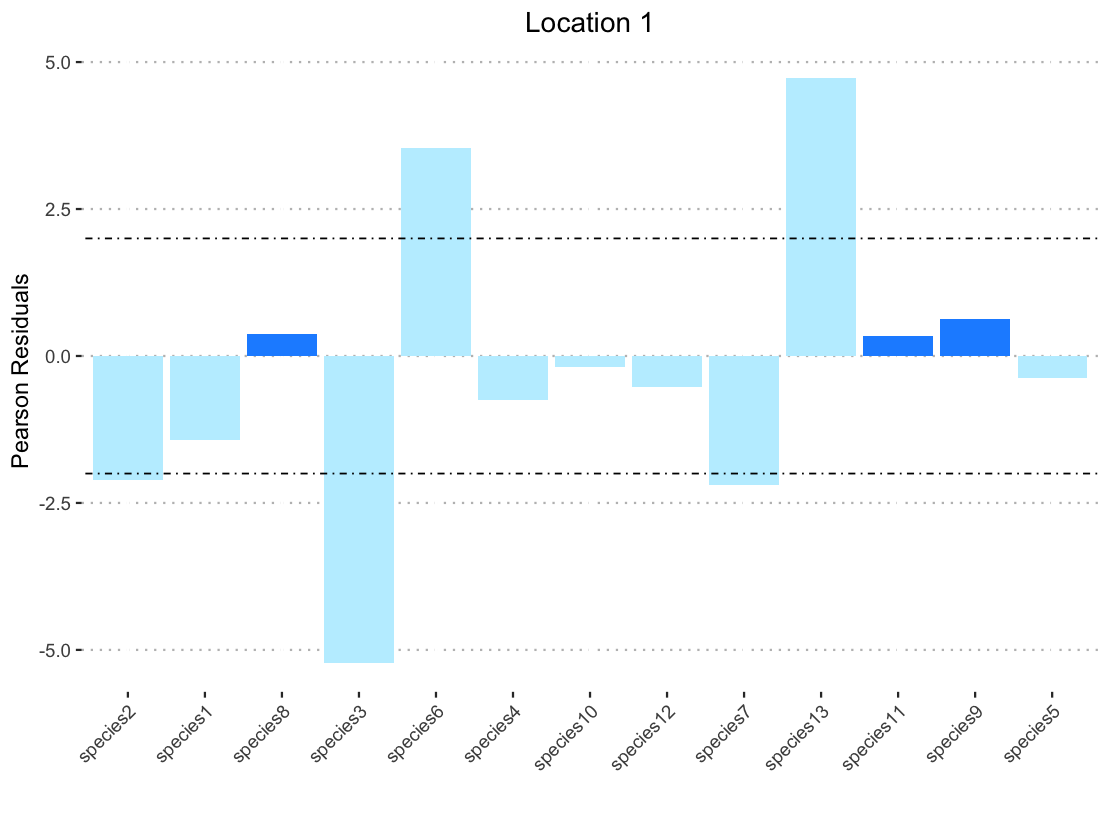
I thought perhaps this was because when you have to specify the dataframe with "data1$color" the filter function was no longer applicable so I broke down by pipe and created a data frame that was pre-filtered to call for the ggplot. But even when this data frame is ordered with arrange the bars are still not the correct color.
test.plot.df2<- data1 %>%
filter(location == "location1") %>%
arrange(species_order)
test.plot.2<- test.plot.df2 %>%
ggplot(aes(
reorder(x = species, species_order),
y= test_residuals,
fill = species))
geom_bar( stat= "identity")
ggtitle("Location 1")
theme_pubclean(
base_size = 14 )
theme(plot.title = element_text(hjust = 0.5),
legend.position = "none")
xlab("") ylab("Pearson Residuals")
scale_x_discrete(guide = guide_axis(angle = 45))
geom_abline(intercept = 2, slope = 0, linetype = "dotdash")
geom_abline(intercept = -2, slope = 0, linetype = "dotdash")
scale_fill_manual(values = test.plot.df2$color)
test.plot.2
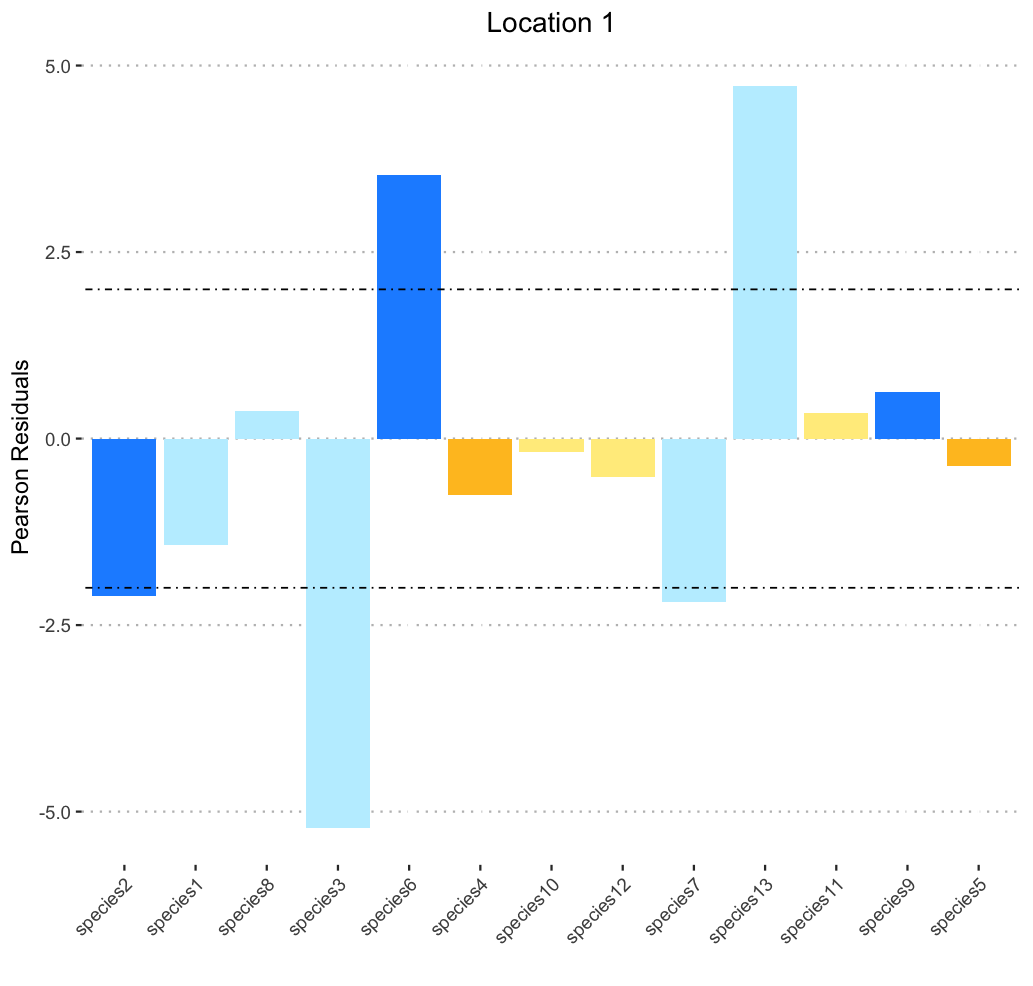
Produces:
I must have a syntax error somewhere, but I cannot seem to find the logic behind the order of column colors produced, and am thus unable to work out how to correct said syntax error. Among (many many) things I have tried, I created a single vector to call for color
test.plot.df2<- data1 %>%
filter(location == "location1") %>%
arrange(species_order)
test_color1<- test.plot.df2$color
test.plot.2<- test.plot.df2 %>%
ggplot(aes(
reorder(x = species, species_order),
y= test_residuals,
fill = species))
geom_bar( stat= "identity")
ggtitle("Location 1")
theme_pubclean(
base_size = 14 )
theme(plot.title = element_text(hjust = 0.5),
legend.position = "none")
xlab("") ylab("Pearson Residuals")
scale_x_discrete(guide = guide_axis(angle = 45))
geom_abline(intercept = 2, slope = 0, linetype = "dotdash")
geom_abline(intercept = -2, slope = 0, linetype = "dotdash")
scale_fill_manual(values = test_color1)
test.plot.2
Which produces the same graph as above. I have also tried creating a new column, with species order as a character, and calling that for fill. This once again produces a miscolored graph:
test.plot.df3<- data1 %>%
filter(location == "location1") %>%
arrange(species_order) %>%
mutate(species_order_character = as.character(species_order))
test.plot.3<- test.plot.df3 %>%
ggplot(aes(
reorder(x = species, species_order),
y= test_residuals,
fill = species_order_character))
geom_bar( stat= "identity")
ggtitle("Location 1")
theme_pubclean(
base_size = 14 )
theme(plot.title = element_text(hjust = 0.5),
legend.position = "none")
xlab("") ylab("Pearson Residuals")
scale_x_discrete(guide = guide_axis(angle = 45))
geom_abline(intercept = 2, slope = 0, linetype = "dotdash")
geom_abline(intercept = -2, slope = 0, linetype = "dotdash")
scale_fill_manual(values = test.plot.df3$color)
test.plot.3
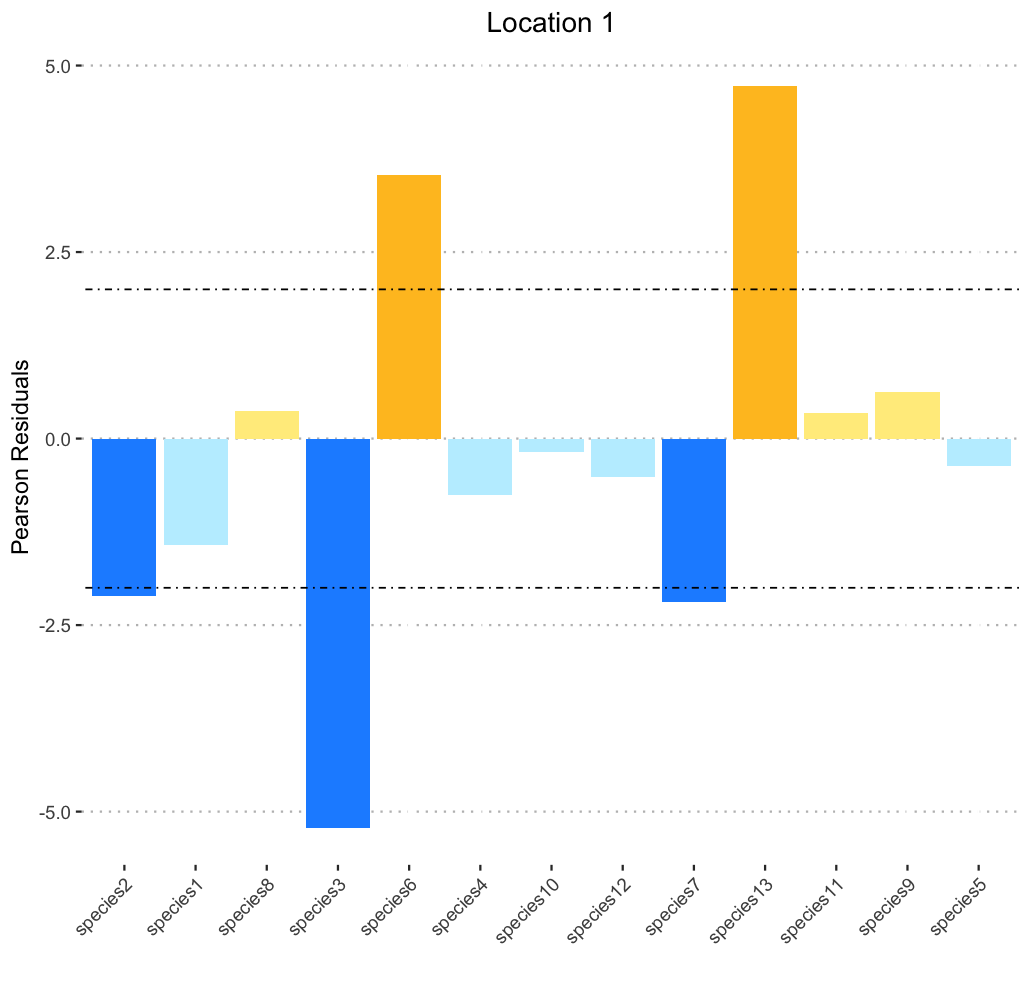
I am at my wits end. I know for each graph I could manually enter the colors like so :
test.plot.4<-data1 %>%
filter(location == "location1") %>%
ggplot(aes(
reorder(x = species, species_order),
y= test_residuals,
fill = color))
geom_bar( stat= "identity")
ggtitle("Location 1")
theme_pubclean(
base_size = 14 )
theme(plot.title = element_text(hjust = 0.5),
legend.position = "none")
xlab("") ylab("Pearson Residuals")
scale_x_discrete(guide = guide_axis(angle = 45))
geom_abline(intercept = 2, slope = 0, linetype = "dotdash")
geom_abline(intercept = -2, slope = 0, linetype = "dotdash")
scale_fill_manual(values = c( "dodgerblue1","goldenrod1", "lightblue1", "lightgoldenrod1"))
test.plot.4
This produces a correctly colored graph, but 1) I would like to have to avoid doing this by hand for each of the many times I have to reproduce this for different locations and different data sets, and 2) even here I can't figure out why the colors need to be ordered that way (ie.: "goldenrod1", "dodgerblue1", "lightgoldenrod1", "lightblue1") to correspond to the correct levels.
Anyone have any insights on what is happening here, and how i might be able to correct my syntax so that I can just call the colors directly from the data frame?
Thanks very much below is the full code to reproduce my data frame :
data1 <- as.data.frame(structure(list(species = c(
"species1", "species1", "species1",
"species1", "species1", "species1", "species2", "species2", "species2",
"species2", "species2", "species2", "species3", "species3", "species3",
"species3", "species3", "species3", "species4", "species4", "species4",
"species4", "species4", "species4", "species5", "species5", "species5",
"species5", "species5", "species5", "species6", "species6", "species6",
"species6", "species6", "species6", "species7", "species7", "species7",
"species7", "species7", "species7", "species8", "species8", "species8",
"species8", "species8", "species8", "species9", "species9", "species9",
"species9", "species9", "species9", "species10", "species10",
"species10", "species10", "species10", "species10", "species11",
"species11", "species11", "species11", "species11", "species11",
"species12", "species12", "species12", "species12", "species12",
"species12", "species13", "species13", "species13", "species13",
"species13", "species13"
), location = c(
"location1", "location2",
"location3", "location4", "location5", "location6", "location1",
"location2", "location3", "location4", "location5", "location6",
"location1", "location2", "location3", "location4", "location5",
"location6", "location1", "location2", "location3", "location4",
"location5", "location6", "location1", "location2", "location3",
"location4", "location5", "location6", "location1", "location2",
"location3", "location4", "location5", "location6", "location1",
"location2", "location3", "location4", "location5", "location6",
"location1", "location2", "location3", "location4", "location5",
"location6", "location1", "location2", "location3", "location4",
"location5", "location6", "location1", "location2", "location3",
"location4", "location5", "location6", "location1", "location2",
"location3", "location4", "location5", "location6", "location1",
"location2", "location3", "location4", "location5", "location6",
"location1", "location2", "location3", "location4", "location5",
"location6"
), test_residuals = c(
-1.43157930150306, -0.314316453493008,
-0.695141335636191, -2.50279485833503, 15.9593244074832, -3.33654341630138,
-2.11214812519871, -0.754659543030408, -2.3490433970076, -1.7153639945355,
19.798140868747, -3.92267054433899, -5.21633871800811, -2.78600907892934,
4.13596459214836, -2.35842831236716, -4.34026196885217, 8.57347502255589,
-0.754659543030408, -2.11214812519871, -1.7153639945355, 9.81355206430024,
-0.0987450246067016, -2.3490433970076, -0.367677794665814, -0.298606543279543,
-0.261519516774949, -0.131369364295332, -0.472983769840402, 0.781602686808182,
3.53010760821268, -5.58101185979998, -5.5626379561955, 5.74088803484089,
-12.2995673766017, 10.0851562256946, -2.18846593288851, -0.161746935435626,
-1.76434843091121, -1.28043017699489, 9.27256034587805, -4.25159798465366,
0.372729803108757, -1.46533093179302, 0.229469416155288, 6.81036162101337,
-2.23476643015094, 0.351490912112304, 0.624530722145124, 1.07723113193857,
-0.262738728590663, -0.945967539680804, 3.3007673589212, -1.36569858688998,
-0.18578433666679, -0.519974923799824, -0.422293423319278, 5.03783441267317,
-0.965694731846794, -0.668900062090651, 0.337429125033733, -0.656846821476658,
-0.250681398015413, -0.153477341599593, -1.30759758387474, 0.686219077483926,
-0.519974923799824, -0.18578433666679, -0.668900062090651, -0.422293423319278,
-0.36984444744839, 1.10535312007138, 4.72231943431065, 0.0138571578271046,
5.16352940820454, -4.08311797265573, -1.90430067033424, 0.0153780833066176
), species_order = c(
2L, 2L, 2L, 2L, 2L, 2L, 1L, 1L, 1L, 1L,
1L, 1L, 4L, 4L, 4L, 4L, 4L, 4L, 6L, 6L, 6L, 6L, 6L, 6L, 13L,
13L, 13L, 13L, 13L, 13L, 5L, 5L, 5L, 5L, 5L, 5L, 9L, 9L, 9L,
9L, 9L, 9L, 3L, 3L, 3L, 3L, 3L, 3L, 12L, 12L, 12L, 12L, 12L,
12L, 7L, 7L, 7L, 7L, 7L, 7L, 11L, 11L, 11L, 11L, 11L, 11L, 8L,
8L, 8L, 8L, 8L, 8L, 10L, 10L, 10L, 10L, 10L, 10L
), color = c(
"lightblue1",
"lightblue1", "lightblue1", "dodgerblue1", "goldenrod1", "dodgerblue1",
"dodgerblue1", "lightblue1", "dodgerblue1", "lightblue1", "goldenrod1",
"dodgerblue1", "dodgerblue1", "dodgerblue1", "goldenrod1", "dodgerblue1",
"dodgerblue1", "goldenrod1", "lightblue1", "dodgerblue1", "lightblue1",
"goldenrod1", "lightblue1", "dodgerblue1", "lightblue1", "lightblue1",
"lightblue1", "lightblue1", "lightblue1", "lightgoldenrod1",
"goldenrod1", "dodgerblue1", "dodgerblue1", "goldenrod1", "dodgerblue1",
"goldenrod1", "dodgerblue1", "lightblue1", "lightblue1", "lightblue1",
"goldenrod1", "dodgerblue1", "lightgoldenrod1", "lightblue1",
"lightgoldenrod1", "goldenrod1", "dodgerblue1", "lightgoldenrod1",
"lightgoldenrod1", "lightgoldenrod1", "lightblue1", "lightblue1",
"goldenrod1", "lightblue1", "lightblue1", "lightblue1", "lightblue1",
"goldenrod1", "lightblue1", "lightblue1", "lightgoldenrod1",
"lightblue1", "lightblue1", "lightblue1", "lightblue1", "lightgoldenrod1",
"lightblue1", "lightblue1", "lightblue1", "lightblue1", "lightblue1",
"lightgoldenrod1", "goldenrod1", "lightgoldenrod1", "goldenrod1",
"dodgerblue1", "lightblue1", "lightgoldenrod1"
)), class = "data.frame", row.names = c(
NA,
-78L
)))
CodePudding user response:
As you've calculated the colour explicitly in your dataframe you can use scale_fill_identity. The only other change is that fill is taken from column color not species. The you get:
test.plot.2<- test.plot.df2 %>%
ggplot(aes(
reorder(x = species, species_order),
y= test_residuals,
fill = color))
geom_bar( stat= "identity")
ggtitle("Location 1")
theme(plot.title = element_text(hjust = 0.5),
legend.position = "none")
xlab("") ylab("Pearson Residuals")
scale_x_discrete(guide = guide_axis(angle = 45))
geom_abline(intercept = 2, slope = 0, linetype = "dotdash")
geom_abline(intercept = -2, slope = 0, linetype = "dotdash")
scale_fill_identity()
test.plot.2
CodePudding user response:
Based on your description, I think you want fill = color instead of fill = species. Then you can construct the values to scale_fill_manual from this vector by setting names.
library(tidyverse)
df <- as.data.frame(structure(list(species = c(
"species1", "species1", "species1",
"species1", "species1", "species1", "species2", "species2", "species2",
"species2", "species2", "species2", "species3", "species3", "species3",
"species3", "species3", "species3", "species4", "species4", "species4",
"species4", "species4", "species4", "species5", "species5", "species5",
"species5", "species5", "species5", "species6", "species6", "species6",
"species6", "species6", "species6", "species7", "species7", "species7",
"species7", "species7", "species7", "species8", "species8", "species8",
"species8", "species8", "species8", "species9", "species9", "species9",
"species9", "species9", "species9", "species10", "species10",
"species10", "species10", "species10", "species10", "species11",
"species11", "species11", "species11", "species11", "species11",
"species12", "species12", "species12", "species12", "species12",
"species12", "species13", "species13", "species13", "species13",
"species13", "species13"
), location = c(
"location1", "location2",
"location3", "location4", "location5", "location6", "location1",
"location2", "location3", "location4", "location5", "location6",
"location1", "location2", "location3", "location4", "location5",
"location6", "location1", "location2", "location3", "location4",
"location5", "location6", "location1", "location2", "location3",
"location4", "location5", "location6", "location1", "location2",
"location3", "location4", "location5", "location6", "location1",
"location2", "location3", "location4", "location5", "location6",
"location1", "location2", "location3", "location4", "location5",
"location6", "location1", "location2", "location3", "location4",
"location5", "location6", "location1", "location2", "location3",
"location4", "location5", "location6", "location1", "location2",
"location3", "location4", "location5", "location6", "location1",
"location2", "location3", "location4", "location5", "location6",
"location1", "location2", "location3", "location4", "location5",
"location6"
), test_residuals = c(
-1.43157930150306, -0.314316453493008,
-0.695141335636191, -2.50279485833503, 15.9593244074832, -3.33654341630138,
-2.11214812519871, -0.754659543030408, -2.3490433970076, -1.7153639945355,
19.798140868747, -3.92267054433899, -5.21633871800811, -2.78600907892934,
4.13596459214836, -2.35842831236716, -4.34026196885217, 8.57347502255589,
-0.754659543030408, -2.11214812519871, -1.7153639945355, 9.81355206430024,
-0.0987450246067016, -2.3490433970076, -0.367677794665814, -0.298606543279543,
-0.261519516774949, -0.131369364295332, -0.472983769840402, 0.781602686808182,
3.53010760821268, -5.58101185979998, -5.5626379561955, 5.74088803484089,
-12.2995673766017, 10.0851562256946, -2.18846593288851, -0.161746935435626,
-1.76434843091121, -1.28043017699489, 9.27256034587805, -4.25159798465366,
0.372729803108757, -1.46533093179302, 0.229469416155288, 6.81036162101337,
-2.23476643015094, 0.351490912112304, 0.624530722145124, 1.07723113193857,
-0.262738728590663, -0.945967539680804, 3.3007673589212, -1.36569858688998,
-0.18578433666679, -0.519974923799824, -0.422293423319278, 5.03783441267317,
-0.965694731846794, -0.668900062090651, 0.337429125033733, -0.656846821476658,
-0.250681398015413, -0.153477341599593, -1.30759758387474, 0.686219077483926,
-0.519974923799824, -0.18578433666679, -0.668900062090651, -0.422293423319278,
-0.36984444744839, 1.10535312007138, 4.72231943431065, 0.0138571578271046,
5.16352940820454, -4.08311797265573, -1.90430067033424, 0.0153780833066176
), species_order = c(
2L, 2L, 2L, 2L, 2L, 2L, 1L, 1L, 1L, 1L,
1L, 1L, 4L, 4L, 4L, 4L, 4L, 4L, 6L, 6L, 6L, 6L, 6L, 6L, 13L,
13L, 13L, 13L, 13L, 13L, 5L, 5L, 5L, 5L, 5L, 5L, 9L, 9L, 9L,
9L, 9L, 9L, 3L, 3L, 3L, 3L, 3L, 3L, 12L, 12L, 12L, 12L, 12L,
12L, 7L, 7L, 7L, 7L, 7L, 7L, 11L, 11L, 11L, 11L, 11L, 11L, 8L,
8L, 8L, 8L, 8L, 8L, 10L, 10L, 10L, 10L, 10L, 10L
), color = c(
"lightblue1",
"lightblue1", "lightblue1", "dodgerblue1", "goldenrod1", "dodgerblue1",
"dodgerblue1", "lightblue1", "dodgerblue1", "lightblue1", "goldenrod1",
"dodgerblue1", "dodgerblue1", "dodgerblue1", "goldenrod1", "dodgerblue1",
"dodgerblue1", "goldenrod1", "lightblue1", "dodgerblue1", "lightblue1",
"goldenrod1", "lightblue1", "dodgerblue1", "lightblue1", "lightblue1",
"lightblue1", "lightblue1", "lightblue1", "lightgoldenrod1",
"goldenrod1", "dodgerblue1", "dodgerblue1", "goldenrod1", "dodgerblue1",
"goldenrod1", "dodgerblue1", "lightblue1", "lightblue1", "lightblue1",
"goldenrod1", "dodgerblue1", "lightgoldenrod1", "lightblue1",
"lightgoldenrod1", "goldenrod1", "dodgerblue1", "lightgoldenrod1",
"lightgoldenrod1", "lightgoldenrod1", "lightblue1", "lightblue1",
"goldenrod1", "lightblue1", "lightblue1", "lightblue1", "lightblue1",
"goldenrod1", "lightblue1", "lightblue1", "lightgoldenrod1",
"lightblue1", "lightblue1", "lightblue1", "lightblue1", "lightgoldenrod1",
"lightblue1", "lightblue1", "lightblue1", "lightblue1", "lightblue1",
"lightgoldenrod1", "goldenrod1", "lightgoldenrod1", "goldenrod1",
"dodgerblue1", "lightblue1", "lightgoldenrod1"
)), class = "data.frame", row.names = c(
NA,
-78L
))) %>%
as_tibble()
df <- df %>%
mutate(color = str_remove(color, "1"))
df %>%
ggplot()
aes(species, test_residuals, fill = color)
scale_fill_manual(values = set_names(unique(df$color), unique(df$color)))
geom_bar(stat = "identity")
facet_wrap(~location, scales = "free")
theme(axis.text.x = element_text(angle = 45))
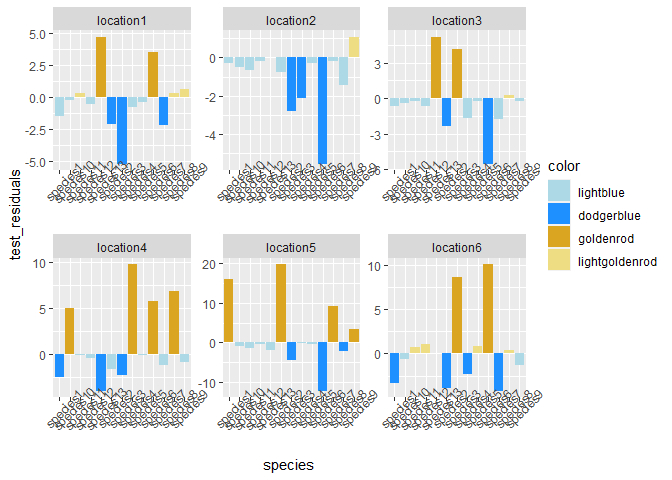
Created on 2022-11-11 with reprex v2.0.2