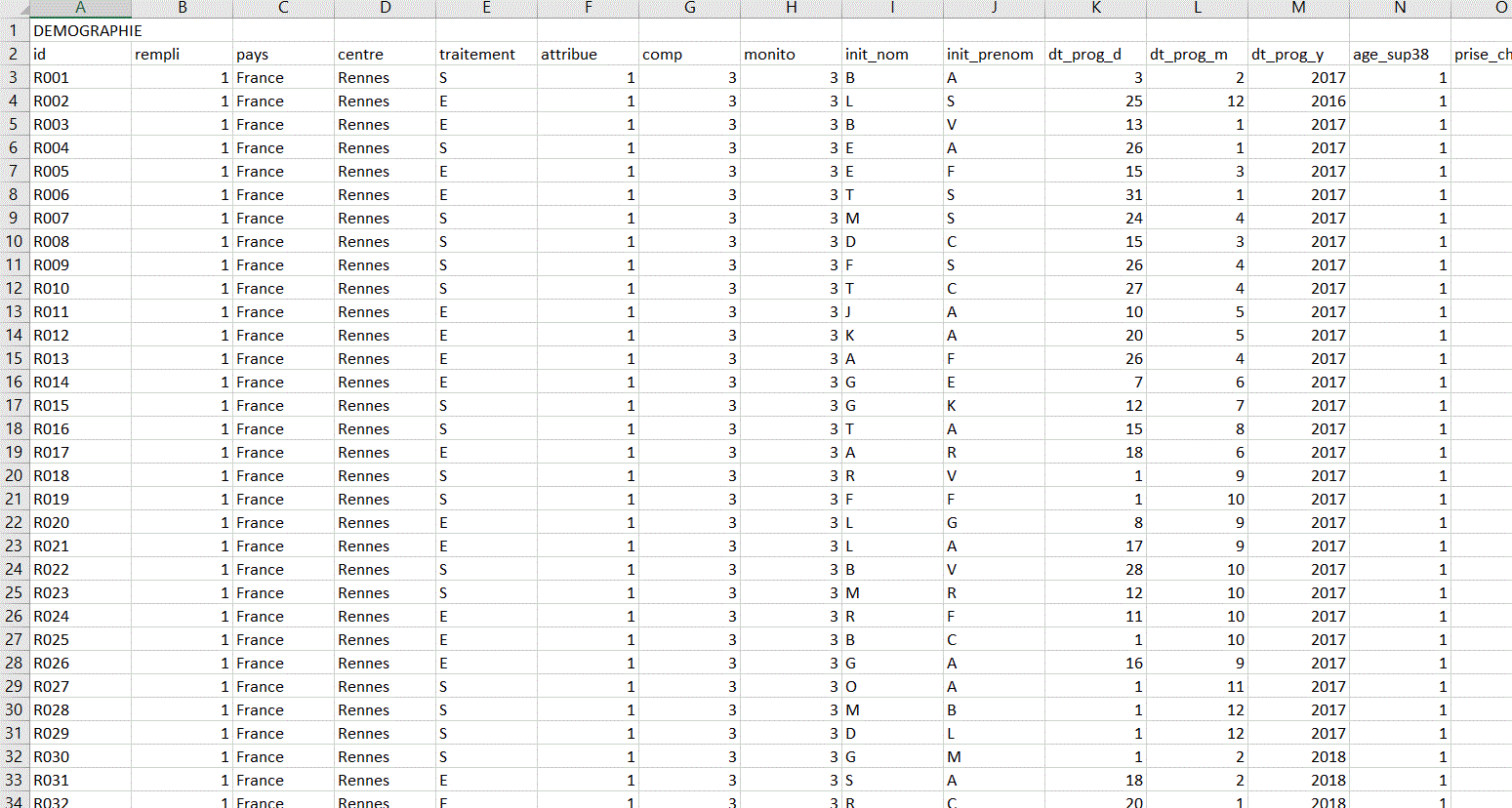
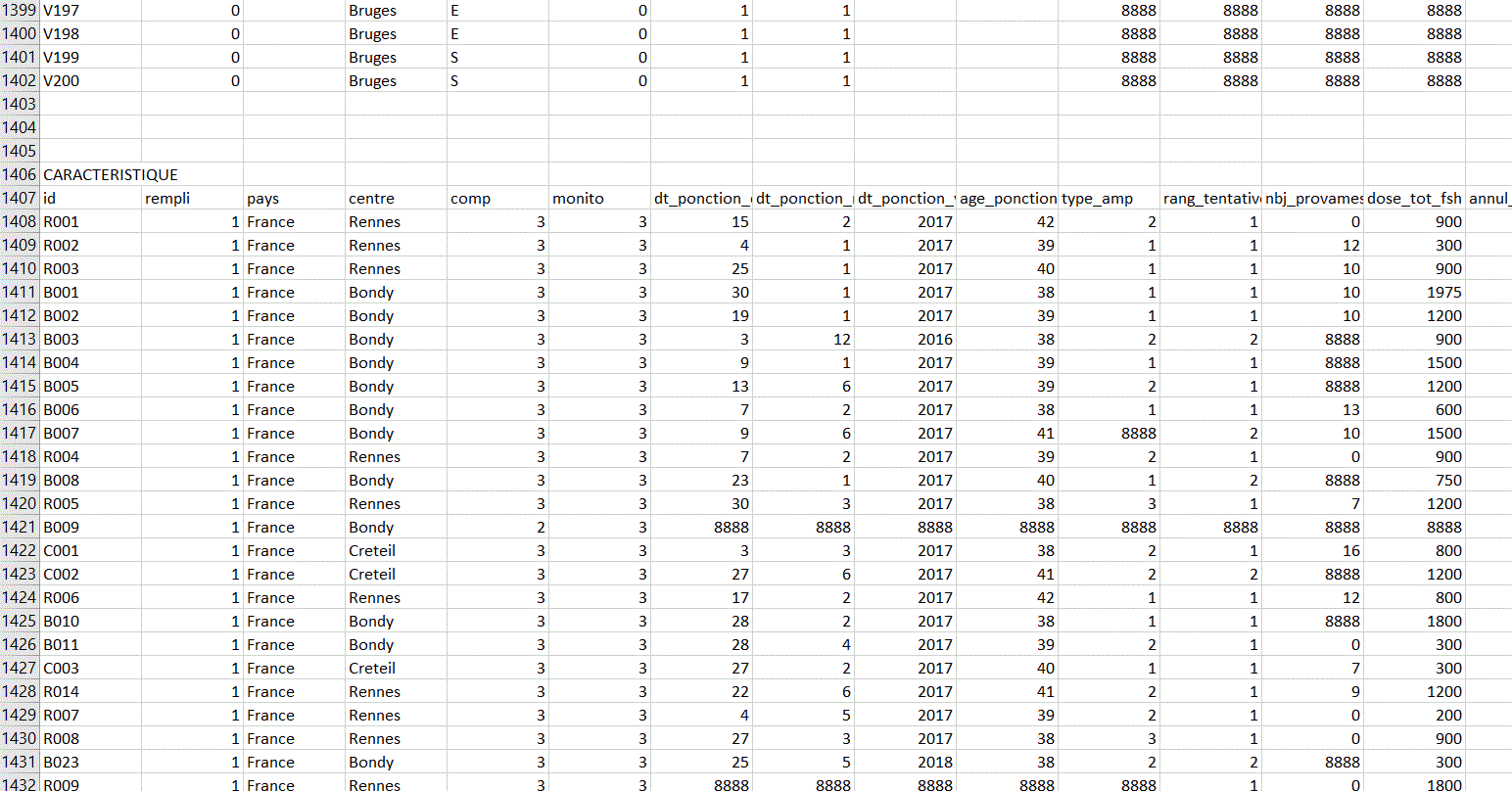
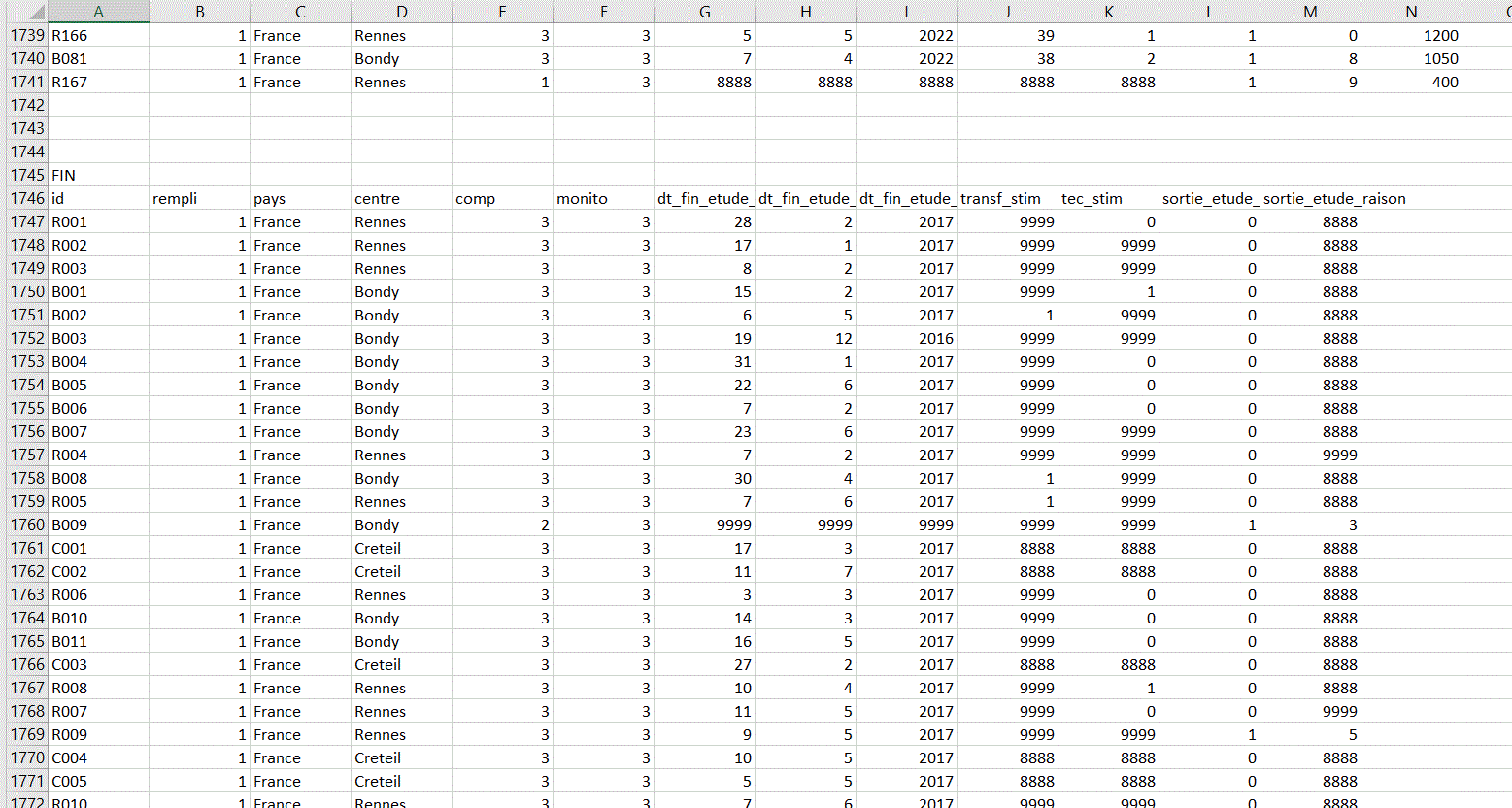
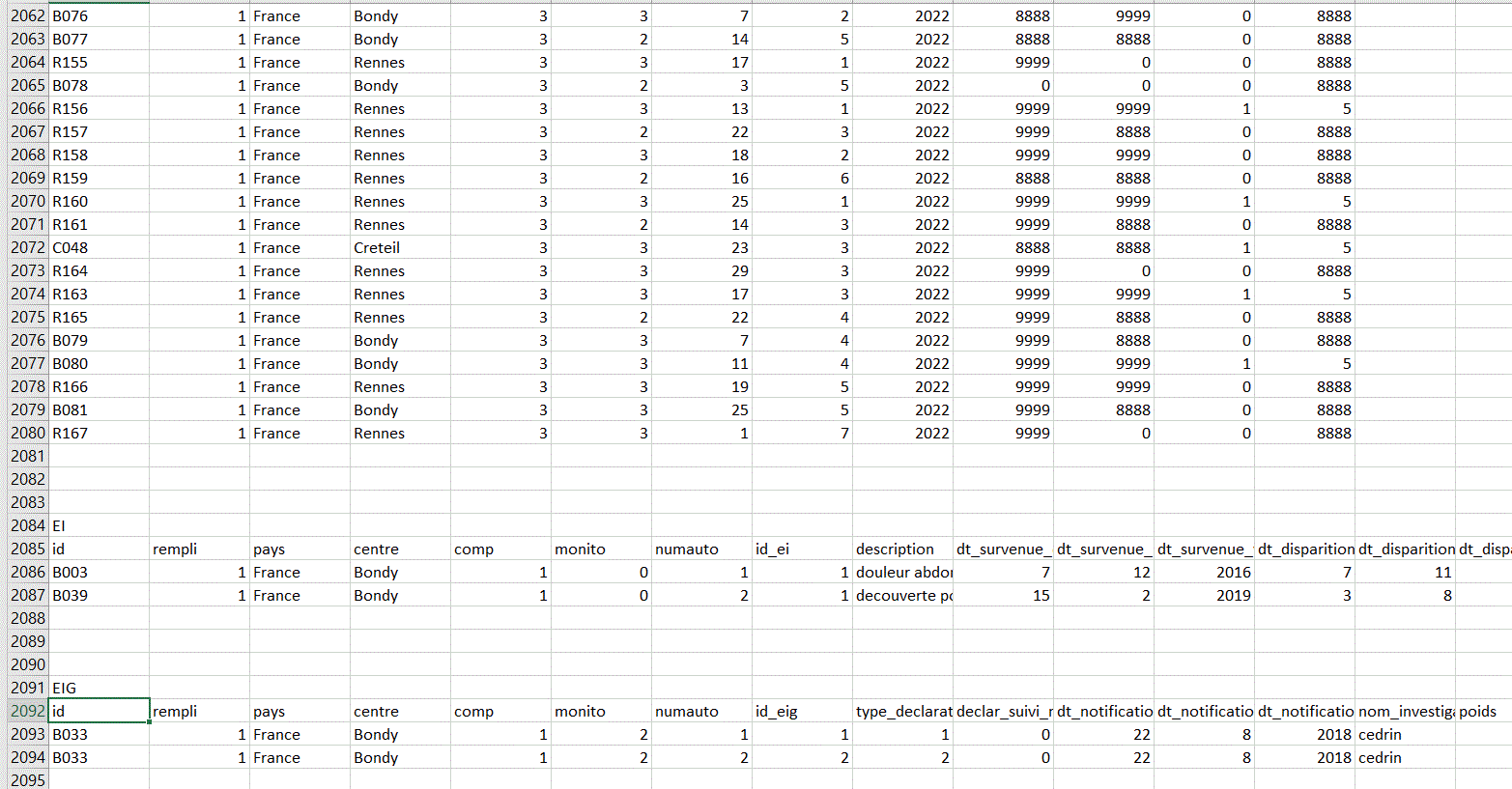
I have 5 databases (DEMOGRAPHIE, CARACTERISTIQUE, FIN, EI ,EIG) all on the same excel sheet. I will have to retrieve each database without having to copy and paste.
This is how the different databases look on the Excel sheet.
I think there are 3 empty rows between each database (but I don't know if this will always be the case).
And there is a row before each dataset corresponding to the title of the dataset. There is a general pattern that indicates where one dataset ends and another begins, but I have no idea how to retrieve each database while keeping the column names in the header
And that is how the datasets appear after importing.
I am wondering if it is more easy to select each after importing or during importing in R
CodePudding user response:
If there are always 3 blank rows separating the tables and they always start with the table name and then column names, the following should work.
Here we first read in the raw data and then parse it to find the positions of each table (using the 3 blank row rule). This is done by finding a match for a string of 3 NA values which is how read_excel() reads blank rows.
For the width, this solution takes advantage of the fact that if you specify the start and end rows, read_excel() can guess the width. Therefore you don't have to figure that out as long as there's nothing else in the table that would confuse it.
For the purposes of demonstration, I made up a minimal sample file with just the first 3 lines of mtcars and iris but it should scale to any arbitrary size.
library(tidyverse)
library(readxl)
library(writexl) # for creating test input
# sample data
d <- structure(list(...1 = c("MTCARS", "mpg", "21", "21", "22.8", NA, NA, NA, "IRIS", "Sepal.Length", "5.0999999999999996", "4.9000000000000004", "4.7"), ...2 = c(NA, "cyl", "6", "6", "4", NA, NA, NA, NA, "Sepal.Width", "3.5", "3", "3.2"), ...3 = c(NA, "disp", "160", "160", "108", NA, NA, NA, NA, "Petal.Length", "1.4", "1.4", "1.3"), ...4 = c(NA, "hp", "110", "110", "93", NA, NA, NA, NA, "Petal.Width", "0.2", "0.2", "0.2"), ...5 = c(NA, "drat", "3.9", "3.9", "3.85", NA, NA, NA, NA, "Species", "setosa", "setosa", "setosa"), ...6 = c(NA, "wt", "2.62", "2.875", "2.3199999999999998", NA, NA, NA, NA, NA, NA, NA, NA), ...7 = c(NA, "qsec", "16.46", "17.02", "18.61", NA, NA, NA, NA, NA, NA, NA, NA), ...8 = c(NA, "vs", "0", "0", "1", NA, NA, NA, NA, NA, NA, NA, NA), ...9 = c(NA, "am", "1", "1", "1", NA, NA, NA, NA, NA, NA, NA, NA), ...10 = c(NA, "gear", "4", "4", "4", NA, NA, NA, NA, NA, NA, NA, NA), ...11 = c(NA, "carb", "4", "4", "1", NA, NA, NA, NA, NA, NA, NA, NA)), class = c("tbl_df", "tbl", "data.frame"), row.names = c(NA, -13L))
d %>% write_xlsx("test.xlsx", format_headers = F, col_names = F)
# read in raw data
raw_input <- read_excel("test.xlsx", col_names = F)
# take first column to find each table
first_col <- raw_input %>% pull(1)
# find positions for each table
table_start_rows <- c(1, which(first_col %in% c(NA,NA,NA))[c(T,F,F)] 3) 1
table_end_rows <- c(which(first_col %in% c(NA,NA,NA))[c(T,F,F)] - 1, length(first_col))
table_lengths <- table_end_rows - table_start_rows
# extract table names
table_names <- first_col[table_start_rows - 1]
# read in each table and capture into list
l <- map2(.x = (table_start_rows - 1),
.y = table_lengths,
.f = ~read_excel("test.xlsx", skip = .x, n_max = .y))
# add table names
names(l) <- table_names
# inspect output
l
#> $MTCARS
#> # A tibble: 3 × 11
#> mpg cyl disp hp drat wt qsec vs am gear carb
#> <chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr>
#> 1 21 6 160 110 3.9 2.62 16.46 0 1 4 4
#> 2 21 6 160 110 3.9 2.875 17.02 0 1 4 4
#> 3 22.8 4 108 93 3.85 2.3199999999999998 18.61 1 1 4 1
#>
#> $IRIS
#> # A tibble: 3 × 5
#> Sepal.Length Sepal.Width Petal.Length Petal.Width Species
#> <chr> <chr> <chr> <chr> <chr>
#> 1 5.0999999999999996 3.5 1.4 0.2 setosa
#> 2 4.9000000000000004 3 1.4 0.2 setosa
#> 3 4.7 3.2 1.3 0.2 setosa
Created on 2022-11-17 with reprex v2.0.2