I am trying to plot a raster in a projected in a coordinated system which follows the curvature of the earth like most projections that are not WGS84. The problem is that the places were the globe wraps around the data should not be plotted outside the globe. I realize that ggplot cannot do a rounded/elliptical plot but how do I mask or remove automatically the data outside the globe? I have to plot more than 100 maps and I can't do this manually especially if I want to change to a different projection.
CodePudding user response:
I would go with one of Robert Hijman's options here, but if you want to create a mask in ggplot, you could do something like this:
library(grid)
y <- seq(0, 1, length = 100)
x <- ifelse(y < 0.5,
-cos(pi/2 * (2 * y - 1)) * 0.125 0.125,
-cos(pi/2 * (2 * y - 1)) * 0.175 0.175)
y <- c(0, y, 1, 0)
x <- c(0, x, 0, 0)
ggplot()
geom_spatraster(data=r)
scale_fill_viridis_c(na.value = 'transparent')
coord_sf(crs = st_crs(" proj=hatano"), expand = FALSE)
annotation_custom(polygonGrob(x = x, y = y,
gp = gpar(col = "white", lwd = 1)))
annotation_custom(polygonGrob(x = 1-x, y = y,
gp = gpar(col = "white", lwd = 1)))
CodePudding user response:
With these data
library(terra)
library(tidyterra)
r1 <- rast('Beck_KG_V1_present_0p5.tif')
r <- subst(r1, 0, NA)
You can do
library(ggplot2)
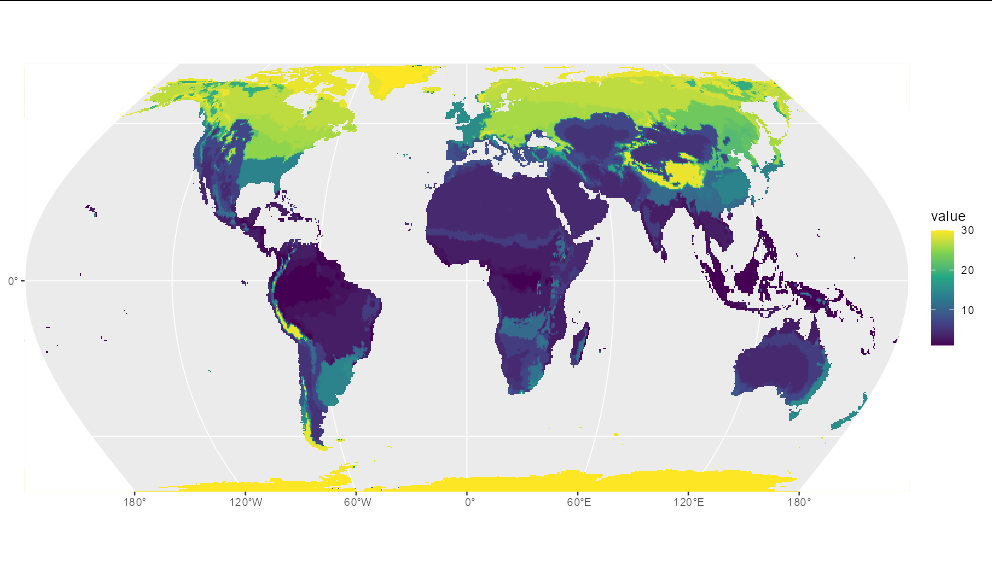
p <- project(r, method="near", " proj=hatano", mask=TRUE)
ggplot() geom_spatraster(data=p) scale_fill_viridis_c(na.value='transparent')
And here are two alternatives with base plot
First with your own color palette and a legend
library(viridis)
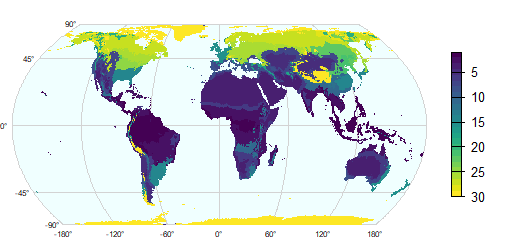
g <- graticule(60, 45, " proj=hatano")
plot(g, background="azure", mar=c(.2,.2,.2,4), lab.cex=0.5, col="light gray")
plot(p, add=TRUE, axes=FALSE, plg=list(shrink=.8), col=viridis(25))
With the colors that came with the file:
coltab(p) <- coltab(r1)
plot(g, background="azure", mar=.5, lab.cex=0.5, col="light gray")
plot(p, add=TRUE, axes=FALSE, col=viridis(25))