I'm trying to use Monte Carlo simulation in order to show how the sum of an uniform sample is normal distributed when the dimension of the sample increase.
More precisely: let define $X ~ U[2,3]$ where $X_1,...,X_n$ is an iid sample from X and $S = \sum_{1}^{n}(X_i). I want use Monte Carlo Simulation in order to show that the distribution of S is approximately normal when n is large (as predicted by Central Limit Theorem).
What I want to show is that when the number of observation in S rise its distribution is more normal. Is also important that I'm talking about the sum of $X_i$, so I'm not considering the general case with the mean.
The problem is that I can obtain a more (or less) normal distribution when I increase (or decrease) the number of time in the Monte Carlo. instead, If I change the sample dimension the differences are VERY low, I can see a normal distribution even when the sample is 10 and, for example, from 10 to 100 i can't notice any significant difference.
Here there is my MWE:
#create random variable with sample size of 1000 that is uniformally distributed
data <- runif(n=10000, min=2, max=3)
hist(data, col='steelblue', main='Histogram from the Uniform')
#I take, for 1000 times, the sum of a sample=10 from X
sample10 <- c()
n = 1000
for (i in 1:n){
sample10[i] = sum(sample(data, 10, replace=TRUE))
}
hist(sample10, col ='steelblue', main='Sample size = 10', prob=TRUE)
qqnorm(sample10); qqline(sample10)
#Increasing the sample dimension
sample100 <- c()
n = 1000
for (i in 1:n){
sample100[i] = sum(sample(data, 100, replace=TRUE))
}
hist(sample100, col ='steelblue', main='Sample size = 100', prob=TRUE)
qqnorm(sample100); qqline(sample100)
What am I doing wrong?
PS. Sorry for my English, any request for clarification is welcome.
CodePudding user response:
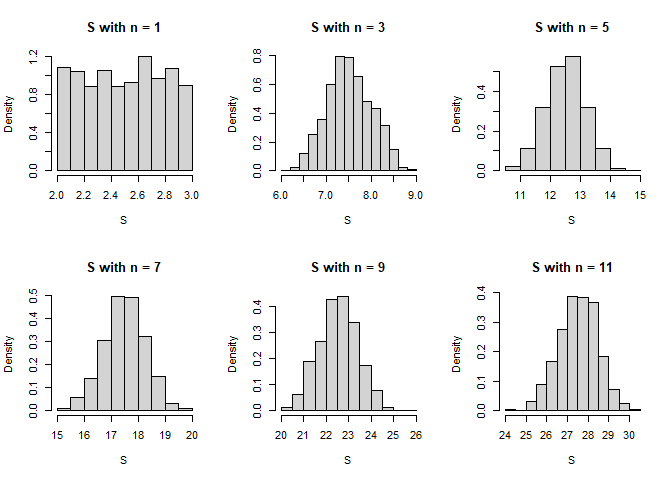
Here is a simulation of the sums of n random uniforms U(2, 3) with n varying from 1 to 11 by steps of 2. Each sum is replicated 1000 times.
set.seed(2022)
nvec <- seq(1, 12, by = 2)
R <- 1e3
S_list <- lapply(nvec, \(n) {
replicate(R, sum(runif(n, 2, 3)))
})
Created on 2022-12-01 with reprex v2.0.2
Now the histograms. You will see that convergence is very quick. That feature is even the basis of a CLT-based pseudo-RNG algorithm for the standard normal.
old_par <- par(mfrow = c(2, 3))
mapply(\(S, n) {
main <- sprintf("S with n = %d", n)
hist(S, main = main, freq = FALSE)
invisible(NULL)
}, S_list, nvec)
#> [[1]]
#> NULL
#>
#> [[2]]
#> NULL
#>
#> [[3]]
#> NULL
#>
#> [[4]]
#> NULL
#>
#> [[5]]
#> NULL
#>
#> [[6]]
#> NULL
par(old_par)
Created on 2022-12-01 with reprex v2.0.2
Don't worry about these NULL's, they are the return value of mapply.
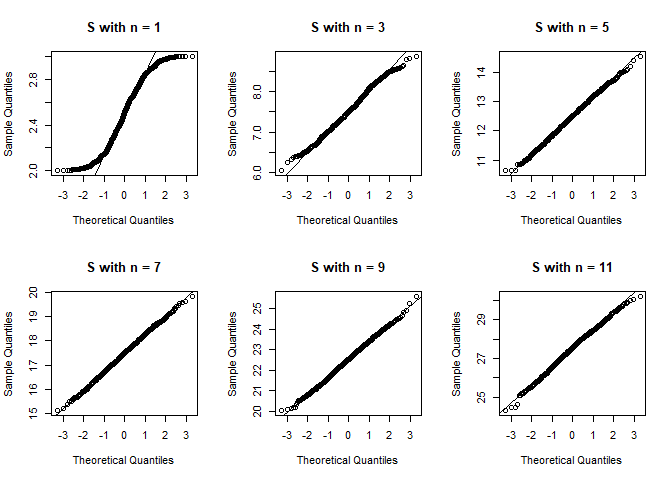
And the QQ-plots.
old_par <- par(mfrow = c(2, 3))
mapply(\(S, n) {
main <- sprintf("S with n = %d", n)
qqnorm(S, main = main)
qqline(S)
}, S_list, nvec)
#> [[1]]
#> NULL
#>
#> [[2]]
#> NULL
#>
#> [[3]]
#> NULL
#>
#> [[4]]
#> NULL
#>
#> [[5]]
#> NULL
#>
#> [[6]]
#> NULL
par(old_par)
Created on 2022-12-01 with reprex v2.0.2
