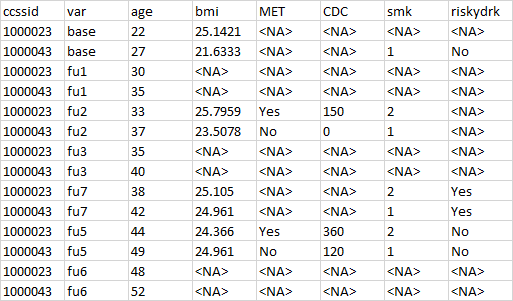
I have this data
dw <- structure(list(ccssid = c(1000023L, 1000043L), base.age = c("22",
"27"), fu1.age = c("30", "35"), fu2.age = c("33", "37"), fu3.age = c("35",
"40"), fu7.age = c("38", "42"), fu5.age = c("44", "49"), fu6.age = c("48",
"52"), base.bmi = c("25.1421", "21.6333"), fu2.bmi = c("25.7959",
"23.5078"), fu7.bmi = c("25.105", "24.961"), fu5.bmi = c("24.366",
"24.961"), fu2.MET = c("150", "0"), fu2.CDC = c("Yes", "No"),
fu5.MET = c("360", "120"), fu5.CDC = c("Yes", "No"), fu6.MET = c(NA_character_,
NA_character_), fu6.CDC = c(NA_character_, NA_character_),
base.smk = c(NA, "1"), fu2.smk = c("2", "1"), fu7.smk = c("2",
"1"), fu5.smk = c("2", "1"), base.riskydrk = c(NA, "No"),
fu7.riskydrk = c("Yes", "Yes"), fu5.riskydrk = c("No", "No"
), base.MET = c(NA, NA), base.CDC = c(NA, NA), fu1.bmi = c(NA,
NA), fu1.MET = c(NA, NA), fu1.CDC = c(NA, NA), fu1.smk = c(NA,
NA), fu1.riskydrk = c(NA, NA), fu2.riskydrk = c(NA, NA),
fu3.bmi = c(NA, NA), fu3.MET = c(NA, NA), fu3.CDC = c(NA,
NA), fu3.smk = c(NA, NA), fu3.riskydrk = c(NA, NA), fu7.MET = c(NA,
NA), fu7.CDC = c(NA, NA), fu6.bmi = c(NA, NA), fu6.smk = c(NA,
NA), fu6.riskydrk = c(NA, NA)), row.names = 1:2, class = "data.frame")
I tried this code below to transform this data, but I am not sure why the values of riskydrk column are switched with smk column in the output. The smk column should have values 1,2, but somehow, it is switched with the values of riskydrk column. Can someone please help me figure out the issue? Thanks!
reshape(dw, direction='long',
varying=c('base.age', 'base.bmi', "base.MET", 'base.CDC', "base.smk", "base.riskydrk",
'fu1.age', 'fu1.bmi', "fu1.MET", 'fu1.CDC', "fu1.smk", "fu1.riskydrk",
'fu2.age', 'fu2.bmi', "fu2.MET", 'fu2.CDC', "fu2.smk", "fu2.riskydrk",
'fu3.age', 'fu3.bmi', "fu3.MET", 'fu3.CDC', "fu3.smk", "fu3.riskydrk",
'fu7.age', 'fu7.bmi', "fu7.MET", 'fu7.CDC', "fu7.smk", "fu7.riskydrk",
'fu5.age', 'fu5.bmi', "fu5.MET", 'fu5.CDC', "fu5.smk", "fu5.riskydrk",
'fu6.age', 'fu6.bmi', "fu6.MET", 'fu6.CDC', "fu6.smk", "fu6.riskydrk"),
timevar='var',
times=c('base', 'fu1', 'fu2', 'fu3', 'fu7', 'fu5', 'fu6'),
v.names=c('age', 'bmi', 'MET', 'CDC', 'smk', 'riskydrk'),
idvar='ccssid')
The desired output should be like this:
CodePudding user response:
I think your problem is, that your times should be suffixes, so reshape is able to guess, which would be way easier.
## make prefixes to suffixes
names(dw) <- strsplit(names(dw), '\\.') |> lapply(rev) |> sapply(paste, collapse='.')
reshape(dw, direction='l', idvar='ccssid', varying=sort(names(dw)[-1]))
# ccssid time age bmi CDC MET riskydrk smk
# 1000023.base 1000023 base 22 25.1421 <NA> <NA> <NA> <NA>
# 1000043.base 1000043 base 27 21.6333 <NA> <NA> No 1
# 1000023.fu1 1000023 fu1 30 <NA> <NA> <NA> <NA> <NA>
# 1000043.fu1 1000043 fu1 35 <NA> <NA> <NA> <NA> <NA>
# 1000023.fu2 1000023 fu2 33 25.7959 Yes 150 <NA> 2
# 1000043.fu2 1000043 fu2 37 23.5078 No 0 <NA> 1
# 1000023.fu3 1000023 fu3 35 <NA> <NA> <NA> <NA> <NA>
# 1000043.fu3 1000043 fu3 40 <NA> <NA> <NA> <NA> <NA>
# 1000023.fu5 1000023 fu5 44 24.366 Yes 360 No 2
# 1000043.fu5 1000043 fu5 49 24.961 No 120 No 1
# 1000023.fu6 1000023 fu6 48 <NA> <NA> <NA> <NA> <NA>
# 1000043.fu6 1000043 fu6 52 <NA> <NA> <NA> <NA> <NA>
# 1000023.fu7 1000023 fu7 38 25.105 <NA> <NA> Yes 2
# 1000043.fu7 1000043 fu7 42 24.961 <NA> <NA> Yes 1
CodePudding user response:
Though the question is not tagged dplyr or tidyr, here is a way with package tidyr.
suppressPackageStartupMessages({
library(tidyr)
})
dw |>
pivot_longer(
cols = -ccssid,
names_to = c("var", ".value"),
names_pattern = "(.*)\\.(.*)"
)
#> # A tibble: 14 × 8
#> ccssid var age bmi MET CDC smk riskydrk
#> <int> <chr> <chr> <chr> <chr> <chr> <chr> <chr>
#> 1 1000023 base 22 25.1421 <NA> <NA> <NA> <NA>
#> 2 1000023 fu1 30 <NA> <NA> <NA> <NA> <NA>
#> 3 1000023 fu2 33 25.7959 150 Yes 2 <NA>
#> 4 1000023 fu3 35 <NA> <NA> <NA> <NA> <NA>
#> 5 1000023 fu7 38 25.105 <NA> <NA> 2 Yes
#> 6 1000023 fu5 44 24.366 360 Yes 2 No
#> 7 1000023 fu6 48 <NA> <NA> <NA> <NA> <NA>
#> 8 1000043 base 27 21.6333 <NA> <NA> 1 No
#> 9 1000043 fu1 35 <NA> <NA> <NA> <NA> <NA>
#> 10 1000043 fu2 37 23.5078 0 No 1 <NA>
#> 11 1000043 fu3 40 <NA> <NA> <NA> <NA> <NA>
#> 12 1000043 fu7 42 24.961 <NA> <NA> 1 Yes
#> 13 1000043 fu5 49 24.961 120 No 1 No
#> 14 1000043 fu6 52 <NA> <NA> <NA> <NA> <NA>
Created on 2022-12-01 with reprex v2.0.2