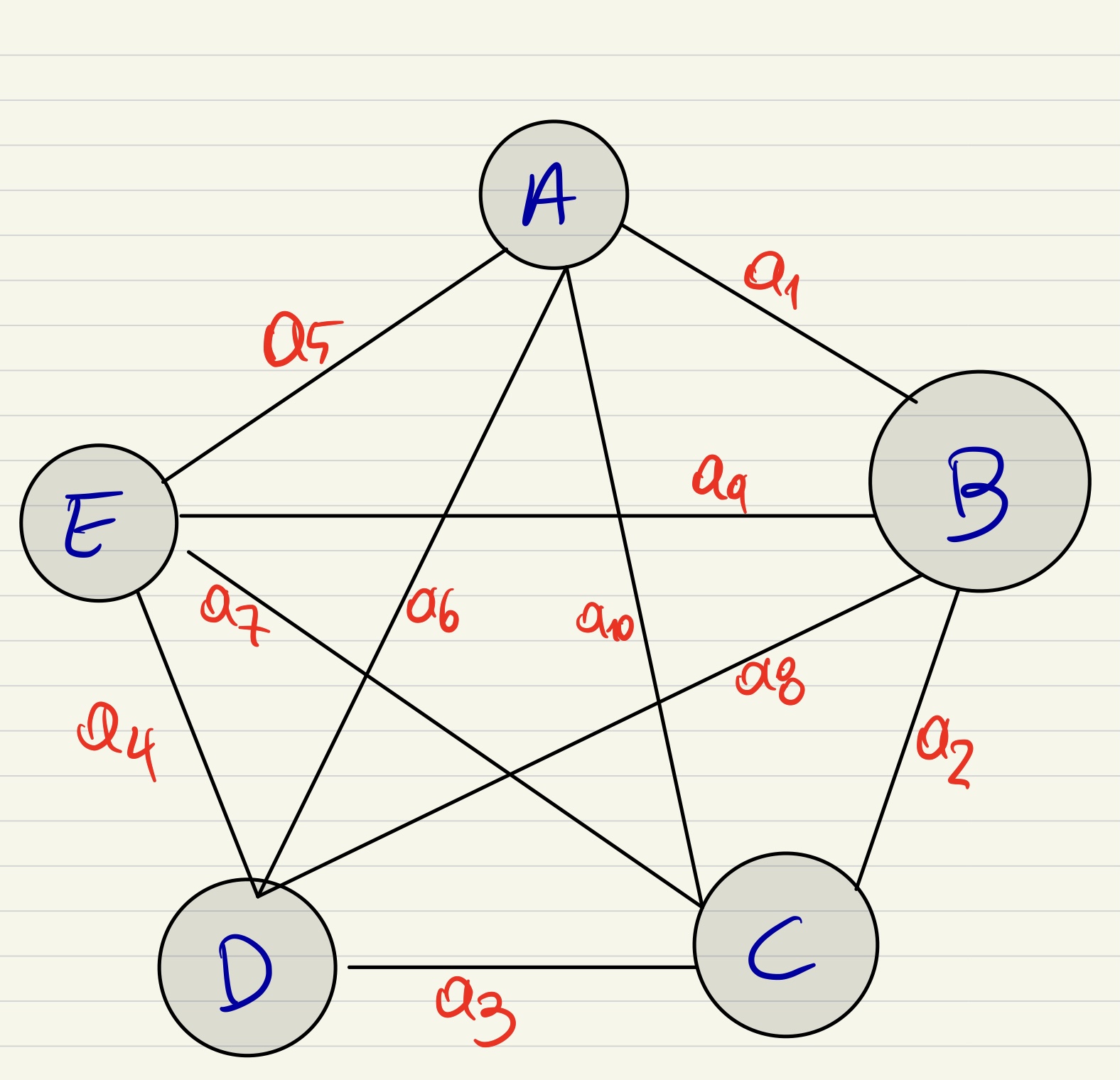
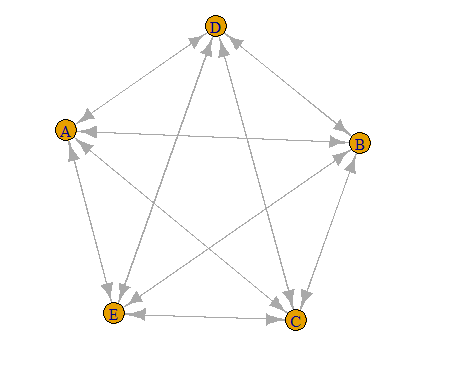
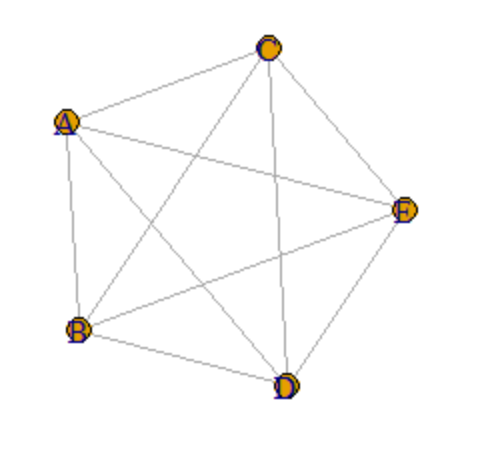
I want to plot an undirected strongly connected graph like the uploaded picture.
I want to use the adjacency matrix in order to plot it.
I tried the igraph package in R :
data <- matrix(c(0,1,0,0,1,
1,0,1,0,0,
0,1,0,1,0,
0,0,1,0,1,
1,0,0,1,0),nrow=5,ncol = 5)
colnames(data) = rownames(data) = LETTERS[1:5]
data
network <- graph_from_adjacency_matrix(data)
plot(network)
But it creates a cycle.
Any help ?
CodePudding user response:
You need only the upper (or lower) triangle of the matrix to be 1, but 0 elsewhere. Make sure you also set mode = "undirected"
library(igraph)
data <- matrix(0, 5, 5, dimnames = list(LETTERS[1:5], LETTERS[1:5]))
data[upper.tri(data)] <- 1
data
#> A B C D E
#> A 0 1 1 1 1
#> B 0 0 1 1 1
#> C 0 0 0 1 1
#> D 0 0 0 0 1
#> E 0 0 0 0 0
network <- graph_from_adjacency_matrix(data, mode = 'undirected')
plot(network)
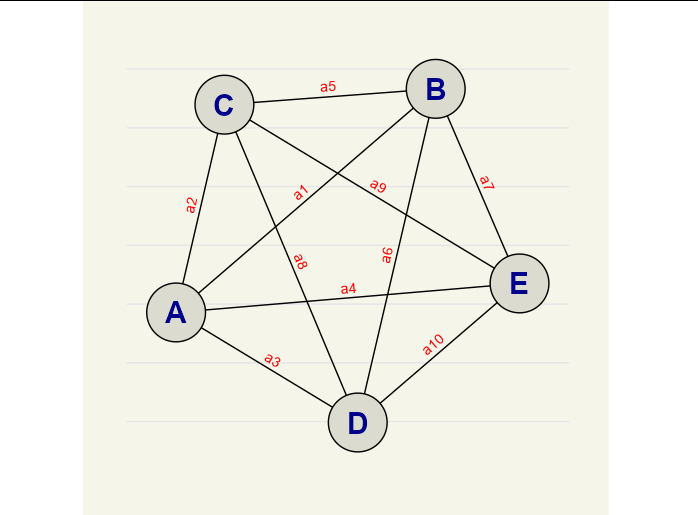
Or, if you want a much more customized appearance (I think your sketch has a nicer aesthetic than the default igraph plot), use ggraph:
library(ggraph)
ggraph(network)
geom_edge_link(aes(label = paste0('a', 1:10)), angle_calc = 'along',
vjust = -0.5, label_colour = 'red')
geom_node_circle(aes(r = 0.1), fill = "#dbdccf")
geom_node_text(aes(label = name), color = 'blue4', fontface = 2,
size = 8)
coord_equal()
theme_graph()
theme(plot.background = element_rect(color = NA, fill = "#f5f6e9"),
panel.grid.major.y = element_line(size = 0.5, color = 'gray90'),
panel.grid.minor.y = element_line(size = 0.5, color = 'gray90'))
CodePudding user response:
You can apply make_full_graph from igraph package, e.g.,
n <- 5
make_full_graph(n) %>%
set_vertex_attr(name = "name", value = head(LETTERS, n)) %>%
plot()
CodePudding user response:
Just modify the matrix as follows:
data <- matrix(c(0,1,1,1,1,
1,0,1,1,1,
1,1,0,1,1,
1,1,1,0,1,
1,1,1,1,0), nrow=5, ncol=5)
Edit
To remove the direction, as Allan Cameron recommended (option mode='undirected'.
network <- graph_from_adjacency_matrix(data, mode='undirected')
plot(network)