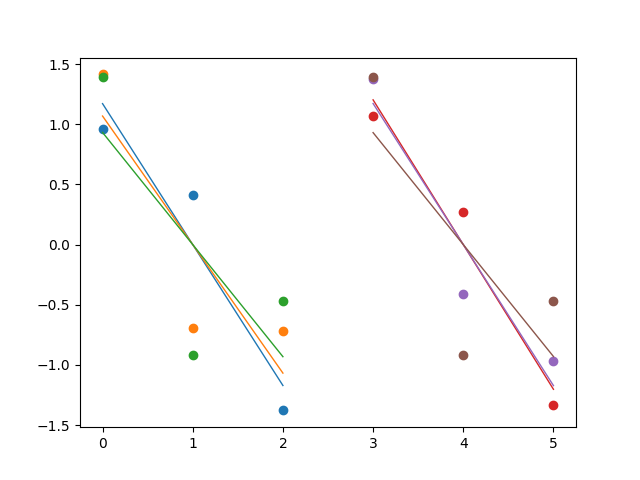
Update with reproducible code:
I have data in form of numeric values that are stored in a nested dictionary called dict_data. This dictionary includes two variables (nested dictionaries), called ple and sampen. To both variables, three rois (lists of data) are attached.
Furthermore, in the nested for loop for variable in variables: for i in range(3):, the dictionary’s data is re-stored into a numpy array for ple and sampen respectively
Next, I apply a quick Z transformation on the numpy array data and save the results in yet another and new numpy array called data_array_z.
Here comes my aim:
I would like to plot a scatterplot for both variables ple and sampen including their three rois(see list rois and the dictionary dict_data). Additionally, I would like to plot linear regression lines across the datapoints in the three rois. An image at the end shows the result that is reproducible with this code. All this works so far.
Problem:
I would like to apply matplotlib’s colormap for both variables’ scatter points and their regression lines. I do not understand how to code the c= or color= commands so that matplotlib plots each new point and regression line with an individual color from a specified colormap, such as tab20c_r.
Currently I receive the following error. AttributeError: 'numpy.ndarray' object has no attribute 'index'
Here is my full code for overview:
import numpy as np
import scipy as sp
import matplotlib.pyplot as plt
from scipy import stats
# Data
rois = ["interoception", "exteroception", "cognitive"]
variables = ["ple", "sampen"]
dict_data = {
"ple":
{"interoception": [-1.10037122285797, -1.12865588460383, -0.703950853686781],
"exteroception": [-1.17398360636007, -1.3990001171012, -1.04528124479161],
"cognitive": [-1.41360171989828, -1.40283540165903, -0.97942989571082]},
"sampen":
{"interoception": [0.949609993391806, 0.855956231042865, 0.960562761361569],
"exteroception": [0.910728054043017, 0.800209085753256, 0.884463613744541],
"cognitive": [0.832925242590743, 0.782835997734383, 0.899277318610028]},
}
# Linear regression for each subject and variable across the three regions
x_pos_1 = np.array([0, 1, 2])
x_pos_2 = np.array([3, 4, 5])
for variable in variables:
for i in range(3):
data_array = np.array([
dict_data[variable]["interoception"][i],
dict_data[variable]["exteroception"][i],
dict_data[variable]["cognitive"][i]
])
# Z normalization
data_array_z = []
for i in data_array:
z = (i - np.mean(data_array)) / np.std(data_array)
data_array_z.append(z)
# Plot
cmap = plt.get_cmap("tab20c_r")
segment_cmap = cmap(np.linspace(0, 1, len(data_array_z)))
if variable == "ple":
reg = sp.stats.linregress(x_pos_1, data_array_z)
plt.scatter(x_pos_1, data_array_z, color=segment_cmap[data_array.index(i)])
plt.plot(x_pos_1, reg[1] reg[0]*x_pos_1,
linestyle="-", linewidth=1, color=segment_cmap[data_array.index(i)],
zorder=2)
elif variable == "sampen":
reg = sp.stats.linregress(x_pos_2, data_array_z)
plt.scatter(x_pos_2, data_array_z)
plt.plot(x_pos_2, reg[1] reg[0]*x_pos_2,
linestyle="-", linewidth=1, zorder=2)
plt.show()
This is what the result looks like when removing the codeline color=segment_cmap[data_array.index(i)] from the first plot, so that matplotlib simply takes its default colormap.
CodePudding user response:
Your code has two problems.
- You reuse the name
iin an inner loop, when the inner loop is completediis now a float, not an index into the columns of your data. - You had this idea of choosing the colors from a qualitative color map, but by default Matplotlib uses colors from a qualitative color map, you have just to use them.
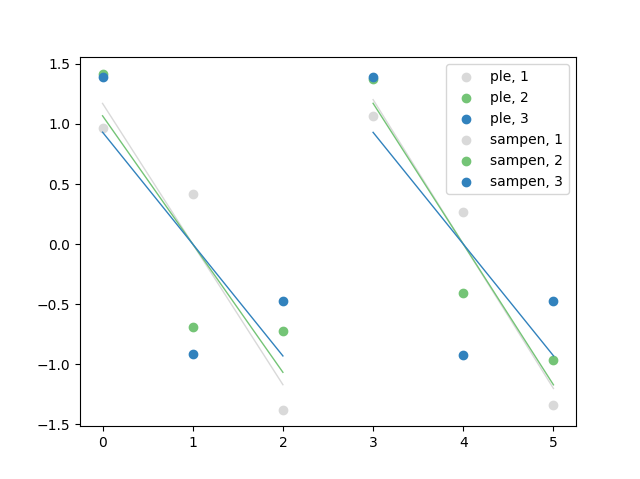
Re ①, I changed the name of the inner loop variable, re ② I used a cycler object to go through the colors from the tab20c_r color map — I used 3 colors, per your code, but the colors are repeated in the plot, is this what you want?
Further, I've added a legend.
import numpy as np
import matplotlib.pyplot as plt
from scipy.stats import linregress
from cycler import cycler
# Data
rois = ["interoception", "exteroception", "cognitive"]
variables = ["ple", "sampen"]
dict_data = {
"ple":
{"interoception": [-1.10037122285797, -1.12865588460383, -0.703950853686781],
"exteroception": [-1.17398360636007, -1.3990001171012, -1.04528124479161],
"cognitive": [-1.41360171989828, -1.40283540165903, -0.97942989571082]},
"sampen":
{"interoception": [0.949609993391806, 0.855956231042865, 0.960562761361569],
"exteroception": [0.910728054043017, 0.800209085753256, 0.884463613744541],
"cognitive": [0.832925242590743, 0.782835997734383, 0.899277318610028]},
}
# Linear regression for each subject and variable across the three regions
x_pos_1 = np.array([0, 1, 2])
x_pos_2 = np.array([3, 4, 5])
plt.gca().set_prop_cycle(
cycler(color=plt.get_cmap("tab20c_r")(np.linspace(0, 1, 3))))
for variable in variables:
for i in range(3):
data_array = np.array([
dict_data[variable]["interoception"][i],
dict_data[variable]["exteroception"][i],
dict_data[variable]["cognitive"][i]
])
# Z normalization
data_array_z = []
for y in data_array:
z = (y - np.mean(data_array)) / np.std(data_array)
data_array_z.append(z)
# Plot
if variable == "ple" or variable == "acw":
reg = linregress(x_pos_1, data_array_z)
print(i)
plt.scatter(x_pos_1, data_array_z, **color, label="%s, %s"%(variable, i 1))
plt.plot(x_pos_1, reg[1] reg[0]*x_pos_1,
linestyle="-", linewidth=1)
elif variable == "sampen":
reg = linregress(x_pos_2, data_array_z)
plt.scatter(x_pos_2, data_array_z, label="%s, %s"%(variable, i 1))
plt.plot(x_pos_2, reg[1] reg[0]*x_pos_2,
linestyle="-", linewidth=1, zorder=2)
plt.legend()
plt.show()