I have data that looks like this:
id Date Time assigned_pat_loc prior_pat_loc Activity
0 45546325 2/7/2011 4:29:38 EIAB^EIAB^6 NaN Admission
1 45546325 2/7/2011 5:18:22 8W^W844^A EIAB^EIAB^6 Observation
2 45546325 2/7/2011 5:18:22 8W^W844^A EIAB^EIAB^6 Transfer to 8W
3 45546325 2/7/2011 6:01:44 8W^W858^A 8W^W844^A Bed Movement
4 45546325 2/7/2011 7:20:44 8W^W844^A 8W^W858^A Bed Movement
5 45546325 2/9/2011 18:36:03 8W^W844^A NaN Discharge-Observation
6 45666555 3/8/2011 20:22:36 EIC^EIC^5 NaN Admission
7 45666555 3/9/2011 1:08:04 53^5314^A EIC^EIC^5 Admission
8 45666555 3/9/2011 1:08:04 53^5314^A EIC^EIC^5 Transfer to 53
9 45666555 3/9/2011 17:03:38 53^5336^A 53^5314^A Bed Movement
I need to find where there were multiple patients (identified with id column) are in the same room at the same time, the start and end times for those, the dates, and room number (assigned_pat_loc). assigned_pat_loc is the current patient location in the hospital, formatted as “unit^room^bed”.
So far I've done the following:
# Read in CSV file and remove bed number from patient location
data = pd.read_csv('raw_data.csv')
data['assigned_pat_loc'] = data['assigned_pat_loc'].str.replace(r"([^^] \^[^^] ).*", r"\1", regex=True)
# Convert Date column to datetime type
patient_data['Date'] = pd.to_datetime(patient_data['Date'])
# Sort dataframe by date
patient_data.sort_values(by=['Date'], inplace = True)
# Identify rows with duplicate room and date assignments, indicating multiple patients shared room
same_room = patient_data.duplicated(subset = ['Date','assigned_pat_loc'])
# Assign duplicates to new dataframe
df_same_rooms = patient_data[same_room]
# Remove duplicate patient ids but keep latest one
no_dups = df_same_rooms.drop_duplicates(subset = ['id'], keep = 'last')
# Group patients in the same rooms at the same times together
df_shuf = pd.concat(group[1] for group in df_same_rooms.groupby(['Date', 'assigned_pat_loc'], sort=False))
And then I'm stuck at this point:
id Date Time assigned_pat_loc prior_pat_loc Activity
599359 42963403 2009-01-01 12:32:25 11M^11MX 4LD^W463^A Transfer
296155 42963484 2009-01-01 16:41:55 11M^11MX EIC^EIC^2 Transfer
1373 42951976 2009-01-01 15:51:09 11M^11MX NaN Discharge
362126 42963293 2009-01-01 4:56:57 11M^11MX EIAB^EIAB^6 Transfer
362125 42963293 2009-01-01 4:56:57 11M^11MX EIAB^EIAB^6 Admission
... ... ... ... ... ... ...
268266 46381369 2011-09-09 18:57:31 54^54X 11M^1138^A Transfer
16209 46390230 2011-09-09 6:19:06 10M^1028 EIAB^EIAB^5 Admission
659699 46391825 2011-09-09 14:28:20 9W^W918 EIAB^EIAB^3 Transfer
659698 46391825 2011-09-09 14:28:20 9W^W918 EIAB^EIAB^3 Admission
268179 46391644 2011-09-09 17:48:53 64^6412 EIE^EIE^3 Admission
Where you can see different patients in the same room at the same time, but I don't know how to extract those intervals of overlap between two different rows for the same room and same times. And then to format it such that the start time and end time are related to the earlier and later times of the transpiring of a shared room between two patients. Below is the desired output.
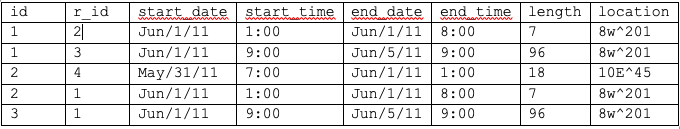
Where r_id is the id of the other patient sharing the same room and length is the number of hours that room was shared.
CodePudding user response:
As suggested, you can use groupby. One more thing you need to take care of is finding the overlapping time. Ideally you'd use datetime which are easy to work with. However you used a different format so we need to convert it first to make the solution easier. Since you did not provide a workable example, I will just write the gist here:
# convert current format to datetime
df['start_datetime'] = pd.to_datetime(df.start_date) df.start_time.astype('timedelta64[h]')
df['end_datetime'] = pd.to_datetime(df.end_date) df.end_time.astype('timedelta64[h]')
df = df.sort_values(['start_datetime', 'end_datetime'], ascending=[True, False])
gb = df.groupby('r_id')
for g, g_df in gb:
g_df['overlap_group'] = (g_df['end_datetime'].cummax().shift() <= g_df['start_datetime']).cumsum()
print(g_df)
This is a tentative example, and you might need to tweak the datetime conversion and some other minor things, but this is the gist.
The cummax() detects where there is an overlap between the intervals, and cumsum() counts the number of overlapping groups, since it's a counter we can use it as a unique identifier.
I used the following threads:
Group rows by overlapping ranges
python/pandas - converting date and hour integers to datetime
Edit
After discussing it with OP the idea is to take each patient's df and sort it by the date of the event. The first one will be the start_time and the last one would be the end_time.
The unification of the time and date are not necessary for detecting the start and end time as they can sort by date and then by the time to get the same order they would have gotten if they did unify the columns. However for the overlap detection it does make life easier when it's in one column.
gb_patient = df.groupby('id')
patients_data_list = []
for patient_id, patient_df in gb_patient:
patient_df = patient_df.sort_values(by=['Date', 'Time'])
patient_data = {
"patient_id": patient_id,
"start_time": patient_df.Date.values[0] patient_df.Time.values[0],
"end_time": patient_df.Date.values[-1] patient_df.Time.values[-1]
}
patients_data_list.append(patient_data)
new_df = pd.DataFrame(patients_data_list)
After that they can use the above code for the overlaps.
