I am fitting the below GAM in mgcv
m3.2 <- bam(pt10 ~
s(year, by = org.type)
s(year, by = region)
s(org.name, bs = 're')
s(org.name, year, bs = 're'),
data = dat,
method = "fREML",
family = betar(link="logit"),
select = T,
discrete = T)
My dat has ~58,000 observations, and the factor org.name has ~2,500 levels - meaning there are a lot of random intercepts and slopes to be fit. Hence, in an attempt to reduce computation time, I have used bam() and the discrete = T option. However, my model has now been running for ~36 hours and still has not fit nor failed and provided me with an error message. I am unsure how long might be reasonable for such a model to take to fit, and therefore how/when to decide to kill the command or not; I don't want to stop the model running if this is normal behaviour/computational time for such a model, but also don't want to waste my time if bam() is stuck going around in circles and will never fit.
Question: How long might be reasonable for such a model to take to fit / what would reasonable computation time for such a model be? Is there a way I can determine if bam() is still making progress or if the command should just be killed to avoid wasting time?
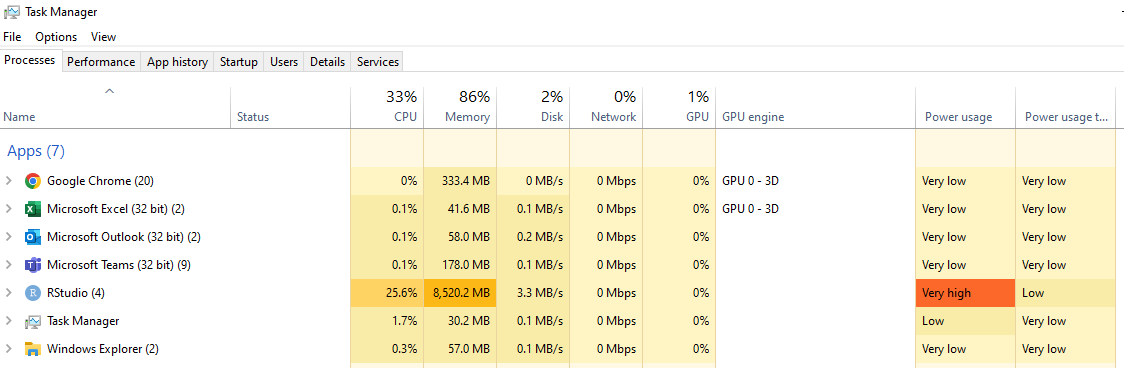
My computer has 16GB RAM and an Intel(R) Core(TM) i7-8565u processor (CPU @ 1.80GHz). In my Windows Task Manager I can see that RStudio is using 20-30% CPU and 20-50% memory and that these usage values are changing and are not static.
CodePudding user response:
To see what bam() is doing, you should have set trace = TRUE in the list passed to the control argument:
ctrl <- gam.control(trace = TRUE)
m3.2 <- bam(pt10 ~
s(year, by = org.type)
s(year, by = region)
s(org.name, bs = 're')
s(org.name, year, bs = 're'),
data = dat,
method = "fREML",
family = betar(link="logit"),
select = TRUE,
discrete = TRUE,
control = ctrl)
That way you'd get printed statements in the console while bam() is doing it's thing. I would check on a much smaller set of data that this actually works (run the example in ?bam() say) within RStudio on Windows; I've never used it so you wouldn't want the trace output to only come once the function had finished, in a single torrent.)
Your problem is the random effects, not really the size of the data; you are estimating 2500 2500 coefficients for your two random effects and another 10 * nlevels(org.type) 10 * nlevels(region) for the two factor by smooths. This is a lot for 58,000 observations.
As you didn't set nthreads it's fitting the model on a single CPU core. I wouldn't necessarily change that though as using 2 threads might just make the memory situation worse. 16GB on Windows isn't very much RAM these days - RStudio is using ~half the available RAM with your other software and Windows using the remaining 36% that Task Manager is reporting is in use.
You should also check if the OS is having to swap memory out to disk; if that's happening then give up as retrieving data from disk for each iteration of model fitting is going to be excruciating, even with a reasonably fast SSD.
The random effects can be done more efficiently in dedicated mixed model software but then you have the problem of writing the GAM bits (the two factor by smooths) in that form - you would write the random effects in the required notation for {brms} or {lme4} respectively as (1 year | org.name) in the relevant part of the formula (or the random argument in gamm4()).
{brms} and {gamm4} can do this bit, but for the former you need to know how to drive {brms} and Stan (which is doing HMC sampling of the posterior), while {lme4}, which is what {gamm4} uses to do the fitting, doesn't have a beta response family. The {bamlss} package has options for this too, but it's quite a complex package so be sure to understand how to specify the model estimation method.
So perhaps revisit your model structure; why do you want a smooth of year for the regions and organisation types, but a linear trend for individual organisations?
