I have an old R code plotting a BMI-plot using Rbase
This is how the old code looks
# BMI-Classes
Gewichtsklassen <- c(0, 18.5, 25, 30, 35, 40, 100)
# nice colors
Farben <- c("skyblue4", "darkgreen", "yellow", "orange", "red", "darkred", "black")
# 100 bodyheights from 1,4m to 2,2m
Koerpergroesse <- seq(1.4, 2.2, length = 100)
# Function to get the BMI class borders
# for any body height
bmi.k <- function(groesse, konstant) {
return(groesse^2 * konstant)
}
# For polygons, we need to give start end enpoint
# so we reverse all body heights
KoerpergroesseREV <- rev(Koerpergroesse)
# calculate all class borders
Klasse1a <- bmi.k(Koerpergroesse, Gewichtsklassen[1])
Klasse1b <- rev(bmi.k(Koerpergroesse, Gewichtsklassen[2]))
Klasse2a <- bmi.k(Koerpergroesse, Gewichtsklassen[2])
Klasse2b <- rev(bmi.k(Koerpergroesse, Gewichtsklassen[3]))
Klasse3a <- bmi.k(Koerpergroesse, Gewichtsklassen[3])
Klasse3b <- rev(bmi.k(Koerpergroesse, Gewichtsklassen[4]))
Klasse4a <- bmi.k(Koerpergroesse, Gewichtsklassen[4])
Klasse4b <- rev(bmi.k(Koerpergroesse, Gewichtsklassen[5]))
Klasse5a <- bmi.k(Koerpergroesse, Gewichtsklassen[5])
Klasse5b <- rev(bmi.k(Koerpergroesse, Gewichtsklassen[6]))
Klasse6a <- bmi.k(Koerpergroesse, Gewichtsklassen[6])
Klasse6b <- rev(bmi.k(Koerpergroesse, Gewichtsklassen[7]))
Klasse7a <- bmi.k(Koerpergroesse, Gewichtsklassen[7])
Klasse7b <- rev(bmi.k(Koerpergroesse, Gewichtsklassen[8]))
#---------------------------------------------------------------------
# lets plot the basic plot
plot(Koerpergroesse, bmi.k(Koerpergroesse, 18), type="n",
xlim=c(1.60, 1.99), ylim=c(40, 125),
xaxt="n", yaxt="n",
cex.axis=1.4, cex.lab=1.3, cex.main=1.7,
xlab="Größe [in m]", ylab="Gewicht [in kg]", main="Body Mass Index")
# add polygons for each class
polygon(c(Koerpergroesse, KoerpergroesseREV), c(Klasse1a, Klasse1b), col=Farben[1])
polygon(c(Koerpergroesse, KoerpergroesseREV), c(Klasse2a, Klasse2b), col=Farben[2])
polygon(c(Koerpergroesse, KoerpergroesseREV), c(Klasse3a, Klasse3b), col=Farben[3])
polygon(c(Koerpergroesse, KoerpergroesseREV), c(Klasse4a, Klasse4b), col=Farben[4])
polygon(c(Koerpergroesse, KoerpergroesseREV), c(Klasse5a, Klasse5b), col=Farben[5])
polygon(c(Koerpergroesse, KoerpergroesseREV), c(Klasse6a, Klasse6b), col=Farben[6])
polygon(c(Koerpergroesse, KoerpergroesseREV), c(Klasse7a, Klasse7b), col=Farben[7])
# add a Gitter
box()
grid(lty="dotdash" ,col="darkgrey")
abline(v=seq(1.65, 1.95, by=0.1), h=seq(50, 110, by=20), lty="dotted", col="grey")
# Legendenbox
legend(x="bottomright", inset=0.005,
legend=c("Untergewicht", "Normalgewicht", "Präadipositas",
"Adipositas Grad I", "Adipositas Grad II", "Adipositas Grad III"),
col=Farben, lwd=6, bg="skyblue")
# X-Achse big ticks
axis(1, at=format(seq(1.60, 2, by=0.1), nsmall=2),
labels=format(seq(1.60, 2, by=0.1), nsmall=2), cex.axis=1.5)
# X-Achse small gray ticks
axis(1, at=seq(1.65, 2, by=0.1), cex.axis=1.2, col.axis="grey")
# Y-Achse Ticks
axis(2, at=seq(40, 120, by=10), cex.axis=1.5)
#---------------------------------------------------------------------
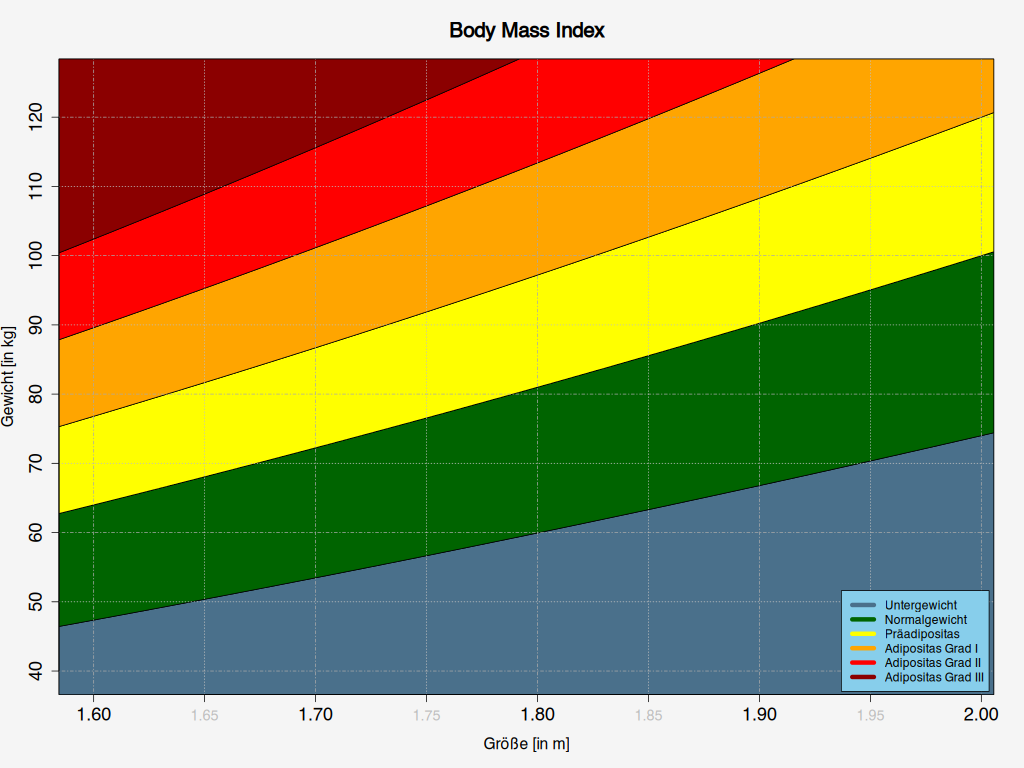
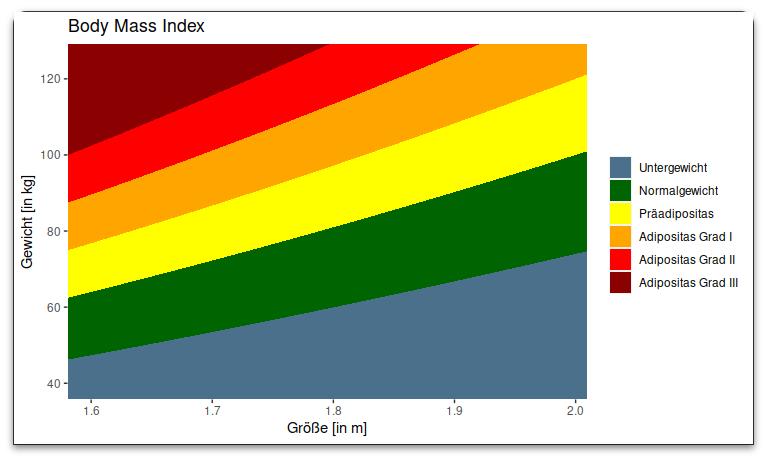
So this works fine and gives me a nice plot like this
So, now I am trying to plot this pic with ggplot2.
I put all data into a data.frame as a "long table"
df1 <- data.frame(Koerpergroesse, Wert=Klasse1a, Class="K1")
df2 <- data.frame(Koerpergroesse, Wert=Klasse2a, Class="K2")
df3 <- data.frame(Koerpergroesse, Wert=Klasse3a, Class="K3")
df4 <- data.frame(Koerpergroesse, Wert=Klasse4a, Class="K4")
df5 <- data.frame(Koerpergroesse, Wert=Klasse5a, Class="K5")
df6 <- data.frame(Koerpergroesse, Wert=Klasse6a, Class="K6")
df7 <- data.frame(Koerpergroesse, Wert=Klasse7a, Class="K7")
df <- rbind(df1, df2, df3, df4, df5, df6, df7 )
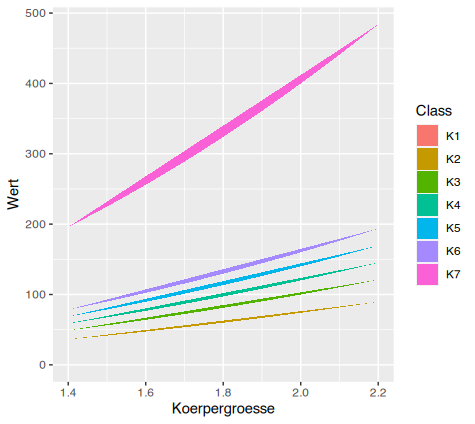
If I call ggplot with geom_polygon like this:
ggplot(df)
aes(x=Koerpergroesse, y=Wert, fill=Class)
geom_polygon()
I get a plot like this:
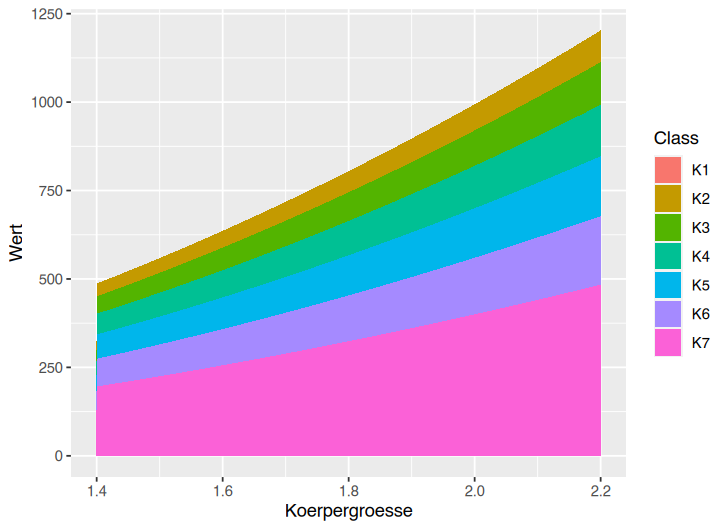
If I use geom_area(), the plot looks even weirder
ggplot(df) aes(x=Koerpergroesse, y=Wert, fill=Class)
geom_area()
The Y-axis seems to be wrong, as K2 for example starts at near 500 and goes up to 1250. These number are not in the dataset:
summary(df)
> summary(df)
Koerpergroesse Wert Class
Min. :1.4 Min. : 0.00 Length:700
1st Qu.:1.6 1st Qu.: 60.83 Class :character
Median :1.8 Median : 91.63 Mode :character
Mean :1.8 Mean :116.95
3rd Qu.:2.0 3rd Qu.:138.66
Max. :2.2 Max. :484.00
So, what am I doing wrong?
How would I plot my BMI plot with ggplot?
EDIT:
Thx for your comments and answers. Although I tagged another answer as the "right one" (because its so simple), the solution to my specific code was to add the "reversed" data to the long table, too
Like this:
### Tidy Data
# bereite "long table" vor
df1 <- data.frame(Gross=c(Koerpergroesse, KoerpergroesseREV), Wert=c(Klasse1a, Klasse1b), Class="K1")
df2 <- data.frame(Gross=c(Koerpergroesse, KoerpergroesseREV), Wert=c(Klasse2a, Klasse2b), Class="K2")
df3 <- data.frame(Gross=c(Koerpergroesse, KoerpergroesseREV), Wert=c(Klasse3a, Klasse3b), Class="K3")
df4 <- data.frame(Gross=c(Koerpergroesse, KoerpergroesseREV), Wert=c(Klasse4a, Klasse4b), Class="K4")
df5 <- data.frame(Gross=c(Koerpergroesse, KoerpergroesseREV), Wert=c(Klasse5a, Klasse5b), Class="K5")
df6 <- data.frame(Gross=c(Koerpergroesse, KoerpergroesseREV), Wert=c(Klasse6a, Klasse6b), Class="K6")
df7 <- data.frame(Gross=c(Koerpergroesse, KoerpergroesseREV), Wert=c(Klasse7a, Klasse7b), Class="K7")
# schreibe alles ins Datenframe df
df <- rbind(df1, df2, df3, df4, df5, df6, df7 )
ggplot(df) aes(x=Gross, y=Wert, fill=Class)
geom_polygon()
coord_cartesian(xlim = c(1.55, 2.05), ylim = c(35, 125), expand = FALSE)
This works and looks as the original plot. (I need to change colors and add a legendbox, but thats no problem)
CodePudding user response:
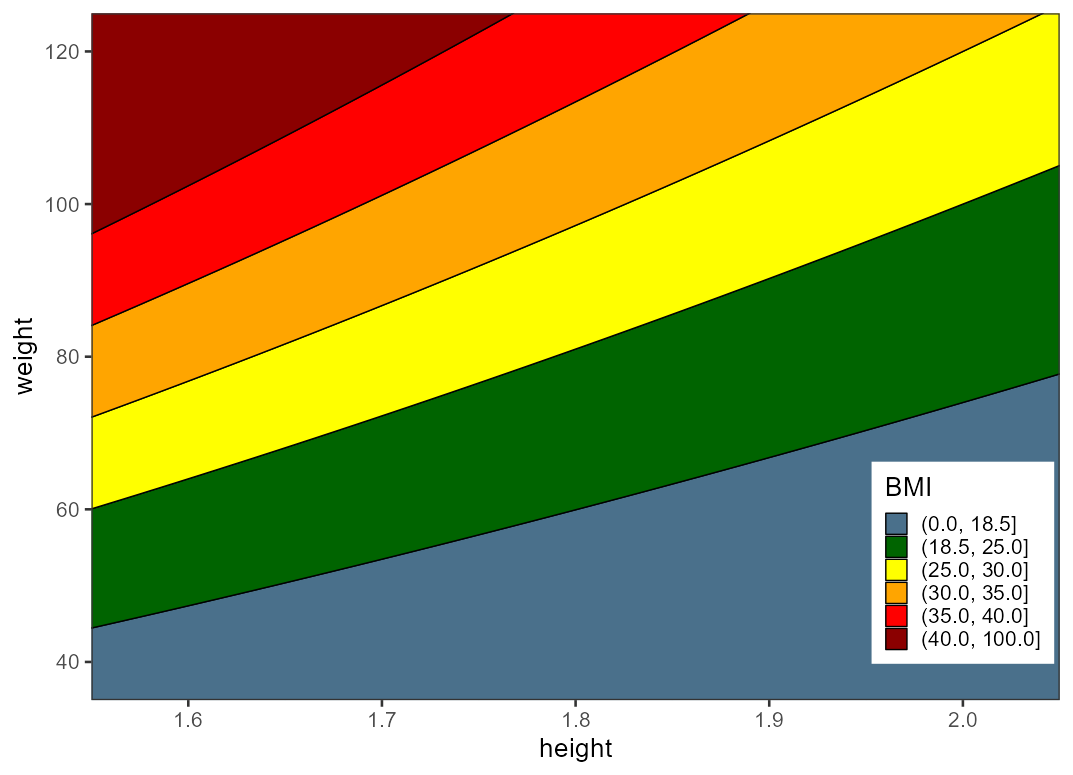
A very simple implementation in ggplot would be to use the BMI formula BMI = weight / height^2 to create a filled contour.
library(ggplot2)
expand.grid(weight = seq(30, 130, 0.1), height = seq(1.5, 2.1, 0.01)) |>
ggplot(aes(height, weight))
geom_contour_filled(aes(z = weight/height^2), color = "black",
breaks = c(0, 18.5, 25, 30, 35, 40, 100))
scale_fill_manual("BMI",
values = rev(c("#8b0000", "red", "#ffa500", "yellow",
"#006400", "#4a708b")))
coord_cartesian(xlim = c(1.55, 2.05), ylim = c(35, 125), expand = FALSE)
theme_bw(base_size = 20)
theme(legend.position = c(0.9, 0.2))
CodePudding user response:
Basic flow of my code:
- For each
K#, identify the nextK#so that we can derive the "bottom" of the polygon; - For each
K#again, interpolate the "other"yvalues from this group'sx/ypair, and then reverse the x/y values; - Combine with the original
df, so that now all ofK1has all ofK1'sKoerpergroessexWert, and then just as many rows withK2'sWertinterpolated values, reversed - I have a
Gewichteframe that provides a good mapping of yourK#to the words you want in the legend ... it's easier to then group usingfill=ClassTxtso that the legend is just... right. For this, I do filter outK7, since ggplot is going to show it even if it is off-screen - Note, if we use
scale_x_continuous(limits=..)(same fory), we get "clipping", meaning we will have incomplete polygons. Instead, we usecoord_cartesianwhere we can have the polygons be continuous with all of its points, even those that are outside of the plot's visible limits.
Gewichte <- data.frame(
ClassTxt = factor(
c("Untergewicht", "Normalgewicht", "Präadipositas", "Adipositas Grad I",
"Adipositas Grad II", "Adipositas Grad III", "UNK"),
levels = c("Untergewicht", "Normalgewicht", "Präadipositas", "Adipositas Grad I",
"Adipositas Grad II", "Adipositas Grad III", "UNK")),
Class = paste0("K", 1:7),
col = Farben
)
# names(Farben) <- paste0("K", 1:7)
slice_head(df, n = 1, by = Class) %>%
transmute(Class, OthClass = lead(Class)) %>%
left_join(df, by = "Class") %>%
mutate(
Wert = if (is.na(OthClass[1])) Inf else {
rev(approx(df$Koerpergroesse[df$Class==OthClass[1]],
df$Wert[df$Class==OthClass[1]],
xout = Koerpergroesse, na.rm = TRUE)$y) },
Koerpergroesse = rev(Koerpergroesse),
.by = Class) %>%
ungroup() %>%
select(-OthClass) %>%
bind_rows(df) %>%
left_join(Gewichte, by = "Class") %>%
filter(Class != "K7") %>%
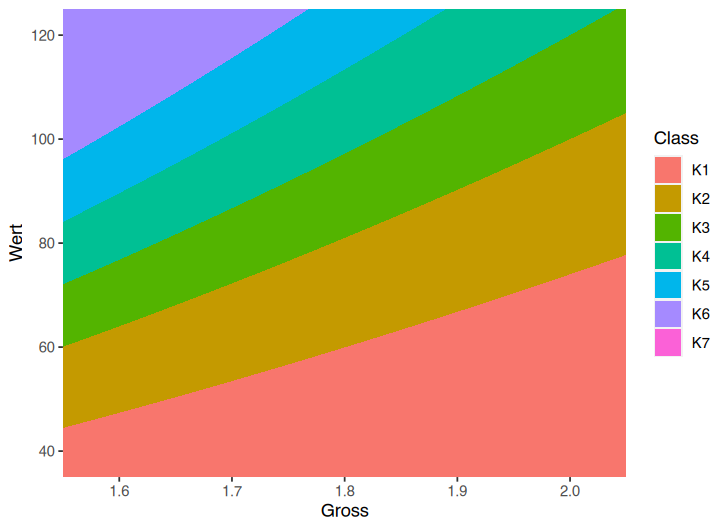
ggplot()
geom_polygon(aes(x = Koerpergroesse, y = Wert, fill = ClassTxt))
coord_cartesian(xlim = c(1.60, 1.99), ylim = c(40, 125))
scale_fill_manual(name = NULL, values = setNames(Gewichte$col, Gewichte$ClassTxt))
labs(title = "Body Mass Index", x = "Größe [in m]", y = "Gewicht [in kg]")
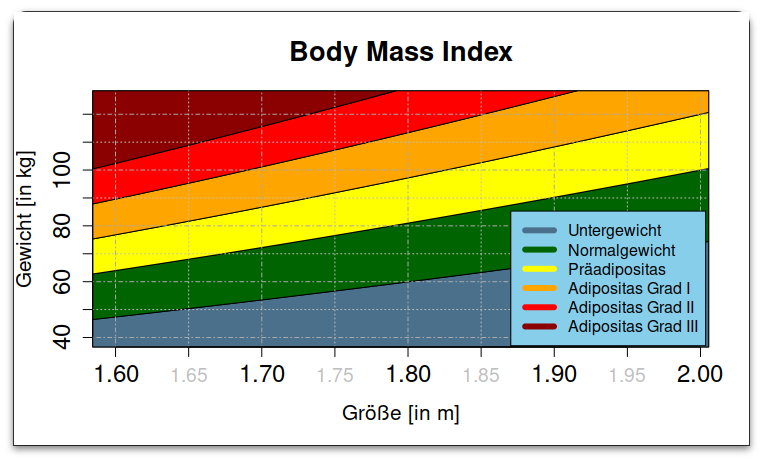
This ggplot output:
The base-graphics you provided produces
(Pretty close.)
I'll leave the rest of the aesthetics (grid, inset legend, axis breaks/labels) to you.