I am trying to calculate a residual matrix (2*ng) for each class of petal length and width, conditioned on sepal length and width for the Iris datset on R(ng = number of observations in the gth class). I need to for each class, plot the residuals as points on a bivariate plot.
I have the following code, but don't feel like this is the right way to go about it. Any help would be appreciated, thanks!
fit = lm(data = iris, Petal.Length Petal.Width ~ Sepal.Length Sepal.Width)
iris$predicted = predict(fit)
iris$residuals = residuals(fit)
iris %>% select(predicted, residuals) %>% head()
ggplot(iris, aes(x=Sepal.Length, y=Petal.Length)) geom_point() geom_point(aes(y=predicted), shape = 1) theme_bw()
ggplot(iris, aes(x=Sepal.Length, y=Petal.Width)) geom_point() geom_point(aes(y=predicted), shape = 1) theme_bw()
ggplot(iris, aes(x=Sepal.Width, y=Petal.Length)) geom_point() geom_point(aes(y=predicted), shape = 1) theme_bw()
ggplot(iris, aes(x=Sepal.Width, y=Petal.Width)) geom_point() geom_point(aes(y=predicted), shape = 1) theme_bw()
CodePudding user response:
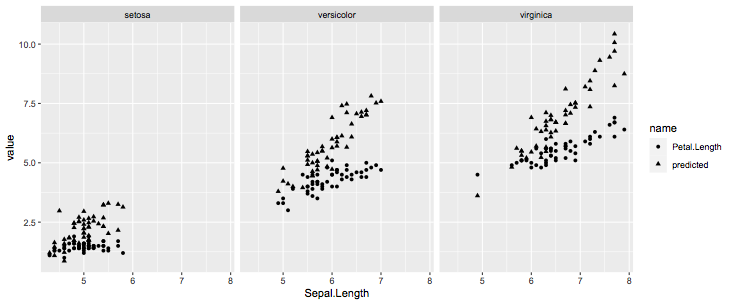
Not very clear from your description and code how you want to plot your residuals. If you want to visualize the predictions and actual values, you just select the variables you need:
library(ggplot2)
library(tidyr)
iris %>% select(Sepal.Length,Species,Petal.Length,predicted) %>%
pivot_longer(-c(Sepal.Length,Species)) %>%
ggplot(aes(x = Sepal.Length,y=value,shape=name))
geom_point()
facet_wrap(~Species)