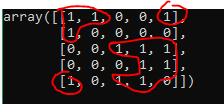
Ideally I'd want the indices of each of the highlighted sections.
CodePudding user response:
That's connected component analysis which has been answered before. Here is a modified possible solution for you.
import numpy as np
from scipy.ndimage.measurements import label
def analysis(array):
labeled, _ = label(array, np.ones((3, 3), dtype=np.int))
for i in np.arange(1, np.max(labeled) 1):
pixels = np.array(np.where(labeled == i))
x1 = np.min(pixels[1, :])
x2 = np.max(pixels[1, :])
y1 = np.min(pixels[0, :])
y2 = np.max(pixels[0, :])
print(str(i) ' | slice: array[' str(y1) ':' str(y2) ', ' str(x1) ':' str(x2) ']')
example1 = np.array([
[1, 1, 0, 0, 1],
[1, 0, 0, 0, 0],
[0, 0, 1, 1, 1],
[0, 0, 0, 1, 1],
[1, 0, 1, 1, 0]
]).astype(bool)
for a in [example1]:
print(a, '\n')
analysis(a)
That's the output (without the example):
[[...]]
1 | slice: array[1:2, 3:5]
2 | slice: array[4:6, 6:8]
3 | slice: array[8:8, 2:2]
CodePudding user response:
teste = ([[1, 1, 0, 0, 1],
[1, 0, 0, 0, 0],
[0, 0, 1, 1, 1],
[0, 0, 0, 1, 1],
[1, 0, 1, 1, 0]])
for x in range(len(teste)):
for y in range(len(teste[x])):
if teste[x][y] == 1:
print('ROW: ' str(x) ' COL: ' str(y) ' IS: ' str(teste[x][y]))
output:
ROW: 0 COL: 0 IS: 1
ROW: 0 COL: 1 IS: 1
ROW: 0 COL: 4 IS: 1
ROW: 1 COL: 0 IS: 1
ROW: 2 COL: 2 IS: 1
ROW: 2 COL: 3 IS: 1
ROW: 2 COL: 4 IS: 1
ROW: 3 COL: 3 IS: 1
ROW: 3 COL: 4 IS: 1
ROW: 4 COL: 0 IS: 1
ROW: 4 COL: 2 IS: 1
ROW: 4 COL: 3 IS: 1

