I have a long dataframe with a datetimeindex that rolls every 5 minutes. However there are gaps and sometimes indexes falling out of the pattern (3 minutes after the previous row, while it should be 5 minutes). I generated an array with datetime values rolling along the same pattern. I would like to modify my dataframe by dropping every row if its index is not in the datetime array, and create an empty row of NaNs for every datetimeindex missing. How could I do that without a for loop ?
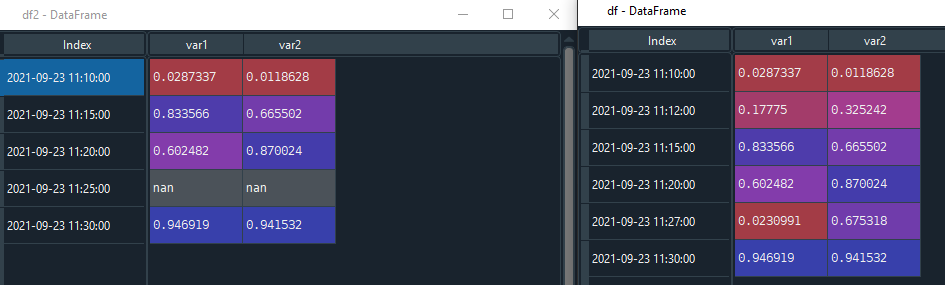
Example code to show what I would like to end up with. df2 represents the corrected df, where the rows with indexes out of the 5min pattern (12min, 27min) dropped, and where the missing 25min row is added and filled with nans.
import pandas as pd
import matplotlib.dates as mdates
import numpy as np
from datetime import datetime
# Create random data
varr1 = np.random.rand(6)
varr2 = np.random.rand(6)
#create the faulty datetimeindex
time_str1 = '23/9/2021 11:10'
date_format_str1 = '%d/%m/%Y %H:%M'
time_str2 = '23/9/2021 11:12'
time_str3 = '23/9/2021 11:15'
time_str4 = '23/9/2021 11:20'
time_str5 = '23/9/2021 11:27'
time_str6 = '23/9/2021 11:30'
# DataFrame with faulty index (some rows do not follow the 5min delta rule, and the 25min mark is missing)
df_time = []
df_time.append(datetime.strptime(time_str1, date_format_str1))
df_time.append(datetime.strptime(time_str2, date_format_str1))
df_time.append(datetime.strptime(time_str3, date_format_str1))
df_time.append(datetime.strptime(time_str4, date_format_str1))
df_time.append(datetime.strptime(time_str5, date_format_str1))
df_time.append(datetime.strptime(time_str6, date_format_str1))
df = pd.DataFrame(index=df_time)
df['var1'] = varr1
df['var2'] = varr2
# Function to generate an array of datetime
def perdelta(start, end, delta):
curr = start
while curr < end:
yield curr
curr = delta
date_arr= []
# Generate datetime that will serve as model
for result in perdelta(df.index[0], df.index[-1], dt.timedelta(minutes=5)):
date_arr.append(result)
date_arr.append(datetime.strptime(time_str6, date_format_str1))
# DataFrame that I want, rows with missing index from datetime array added and filled with NaNs, and the ones falling out of the 5min pattern dropped
df2 = pd.DataFrame({'var1': [varr1[0], varr1[2], varr1[3], np.nan, varr1[-1]], 'var2': [varr2[0], varr2[2], varr2[3], np.nan, varr2[-1]]},index=date_arr)
CodePudding user response:
You can try the example code bellow.
import pandas as pd
import numpy as np
from datetime import timedelta, datetime
varr1 = np.random.rand(6)
varr2 = np.random.rand(6)
date_format = '%d/%m/%Y %H:%M'
dates = ['23/9/2021 11:10',
'23/9/2021 11:12',
'23/9/2021 11:15',
'23/9/2021 11:20',
'23/9/2021 11:27',
'23/9/2021 11:30']
indexes = [datetime.strptime(date, date_format) for date in dates]
df = pd.DataFrame({'var1': varr1, 'var2': varr2}, index=indexes)
expected_dates = []
cur = indexes[0]
end = indexes[-1]
while cur <= end:
expected_dates.append(cur)
cur = timedelta(minutes=5)
items = []
for index in expected_dates:
if index in df.index:
item = {
'var1': df.loc[index, 'var1'],
'var2': df.loc[index, 'var2'],
}
else:
item = {
'var1': np.nan,
'var2': np.nan
}
items.append(item)
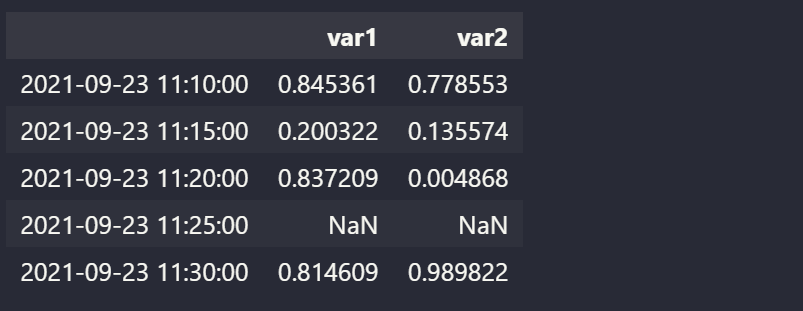
df2 = pd.DataFrame(items, index=expected_dates)
df2
This is ouput