Our analyst has performed a propensity score analysis on our data. Basically, he used country, age and biological start year to "balance" the female and male population in our dataset. He has done an overlap assessment between the two groups (female & male) and looked at the linearized propensity score to see if there is "good" overlap.
Dataset:
structure(list(gender = c(0, 1, 0, 0, 1, 1, 1, 1, 0, 0, 1, 0,
1, 0, 1, 1, 1, 0, 0, 1), country = structure(c(1L, 2L, 2L, 3L,
1L, 1L, 1L, 1L, 1L, 1L, 3L, 3L, 3L, 1L, 1L, 1L, 1L, 1L, 1L, 1L
), .Label = c("CH", "CZ", "DK", "IS", "NL", "NO", "PT", "RO",
"SE", "SF", "SI", "TR", "UK"), class = "factor"), age = c(39,
37, 54, 33, 30, 62, 30, 48, 34, 40, 39, 41, 29, 31, 37, 27, 22,
23, 21, 31), bio_drug_name = structure(c(1L, 1L, 4L, 3L, 1L,
3L, 4L, 3L, 1L, 4L, 3L, 5L, 4L, 4L, 1L, 5L, 1L, 3L, 4L, 2L), .Label = c("adalimumab",
"certolizumab", "etanercept", "golimumab", "infliximab"), class = "factor"),
bio_drug_start_year = c(2007, 2011, 2012, 2012, 2012, 2004,
2012, 2012, 2012, 2012, 2012, 2012, 2016, 2015, 2013, 2015,
2013, 2013, 2014, 2013), asdas_crp_cii_6month = c(1, 1, 0,
1, 0, 0, 1, 0, 1, 1, 0, 1, 1, 1, 0, 1, 1, 0, 1, 0), bio_drug_start_year_centered = c(-8,
-4, -3, -3, -3, -11, -3, -3, -3, -3, -3, -3, 1, 0, -2, 0,
-2, -2, -1, -2), age_std = structure(c(-0.211016383746095,
-0.375088510873223, 1.01952456970737, -0.70323276512748,
-0.949340955818173, 1.67581307821588, -0.949340955818173,
0.527308188325984, -0.621196701563916, -0.12898032018253,
-0.211016383746095, -0.046944256618966, -1.03137701938174,
-0.867304892254609, -0.375088510873223, -1.19544914650887,
-1.60562946432669, -1.52359340076312, -1.68766552789025,
-0.867304892254609), .Dim = c(20L, 1L)), ID = 1:20), na.action = structure(c(`111395` = 169L,
`769107` = 2619L, `844107` = 2624L, `164325` = 2681L, `1011013` = 2728L,
`114174` = 2763L, `116484` = 2778L, `231118` = 3058L), class = "omit"), row.names = c("463",
"7729", "7756", "8306", "8324", "128", "8440", "8450", "8663",
"8809", "8840", "8857", "9020", "9033", "9101", "9324", "9377",
"9523", "9702", "9718"), class = "data.frame")
Code used to create PS-model and calculate linearized PS-score for male and females
psmod = glm( gender ~ country age_std bio_drug_start_year_centered, family = 'binomial', data = dat)
psmod = step(psmod, scope = list(lower = ~country age_std bio_drug_start_year_centered,
upper = ~(country age_std bio_drug_start_year_centered)^2
poly(dat$age_std,degree=3)[,2] poly(dat$age_std,degree=3)[,3]
poly(dat$bio_drug_start_year_centered,degree=3)[,2]
poly(dat$bio_drug_start_year_centered,degree=3)[,3]
),
direction='forward' )
summary(psmod)
# Predict ps-score
ps = predict(psmod, type= 'response')
lps = log(ps/(1-ps))
# Overlap assessment
par(mfrow=c(2,1))
min.lps = min(lps)
max.lps = max(lps)
hist(lps[dat$gender==0], breaks=50,main='male', xlab='Linearized ps-score', xlim=c(min.lps,max.lps))
hist(lps[dat$gender==1], breaks=50,main='female', xlab='Linearized ps-score', xlim=c(min.lps,max.lps))
Here is the output of the image
Although this is fine for him, it is not sufficient for a scientific journal. I would like to use ggplot to create a nice histogram and show overlap between the males and females. There are some nice examples over here However, since the lenghts of the linearized PS-scores differ, I am not sure how to turn this into a dataset and then use it on ggplot.
CodePudding user response:
I couldn't get most of the provided code to run, but if the issue is that the two variables you want to populate histograms with have different numbers of values then something like the following should work:
library(tidyverse)
score_a <- rnorm(n = 50, mean = 0, sd = 1)
score_b <- rnorm(n = 75, mean = 2, sd = 0.75)
# Basic plot:
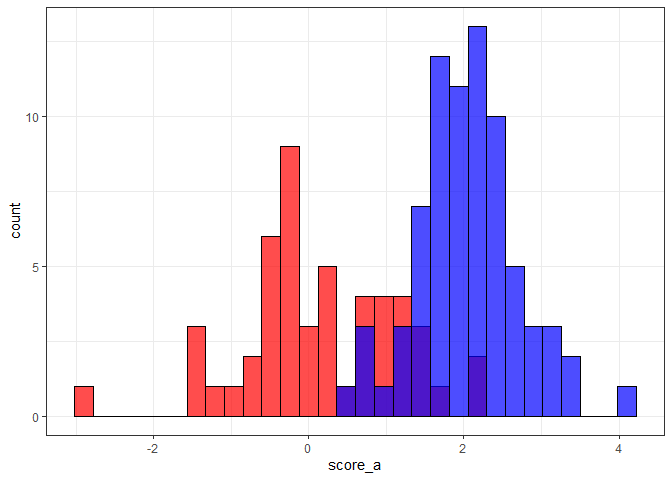
ggplot()
# Add one histogram:
geom_histogram(aes(score_a), color = "black", fill = "red", alpha = 0.7)
# Add second, which has a different number of values
geom_histogram(aes(score_b), color = "black", fill = "blue", alpha = 0.7)
# Black and white theme
theme_bw()
#> `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
#> `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
Edit: If you want to have more control over the x-axis and set it based on min/max of your values, it could look something like the below example. Note that here I've used the round() function because of the values I'm using for the example, but you could omit this and labels = or breaks = seq(from = min_x, to = max_x, by = 0.5) instead if rounding isn't necessary.
# Labeling the x-axis based on the min/max might look like this:
# Define axis breaks & labels:
min_x <- min(c(score_a, score_b))
max_x <- max(c(score_a, score_b))
ggplot()
# Add one histogram:
geom_histogram(aes(score_a), color = "black", fill = "red", alpha = 0.7)
# Add second, which has a different number of values
geom_histogram(aes(score_b), color = "black", fill = "blue", alpha = 0.7)
# Black and white theme
theme_bw()
scale_x_continuous(
breaks = round(x = seq(from = min_x, to = max_x, by = 0.5),
digits = 1),
labels = round(x = seq(from = min_x, to = max_x, by = 0.5),
digits = 1))
#> `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
#> `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
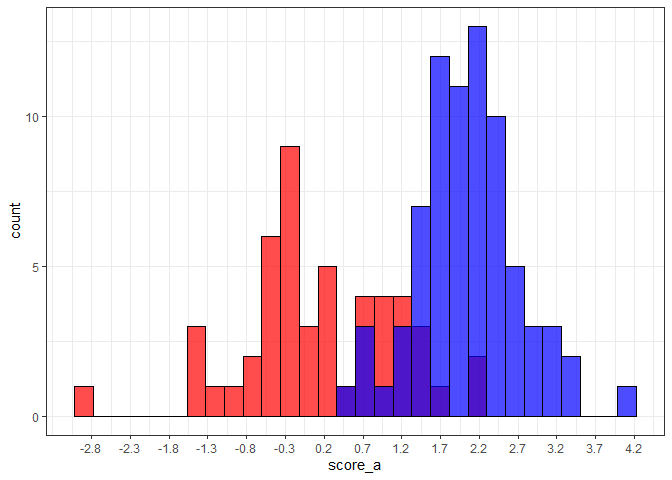
Created on 2021-09-24 by the reprex package (v2.0.0)
CodePudding user response:
Here is one solution. bind lps to the original data and then use ggplot2 to plot and fill color with gender.
dat2 <- cbind(dat, lps)
library(ggplot2)
library(dplyr)
dat2 <- mutate(dat2, gender = as.character(gender))
ggplot(dat2)
geom_histogram(aes(x= lps, fill = gender), bins = 10)
