I have a quite huge dataframe (nearly 100,000 observations with about 40 variables) from which I want ggplot to draw scatterplots with lm- or loess-lines. But the lines should be calculated only based on a certain quantile of variable-values of each observation date. And I would like to do the filtering or subsetting directly in ggplot without creating a new data object or subdataframe in advance.
As my 'real' dataframe would be too large I created fictive example with a dataframe of 144 observations named df_Bandvals (Code at the end of the post).
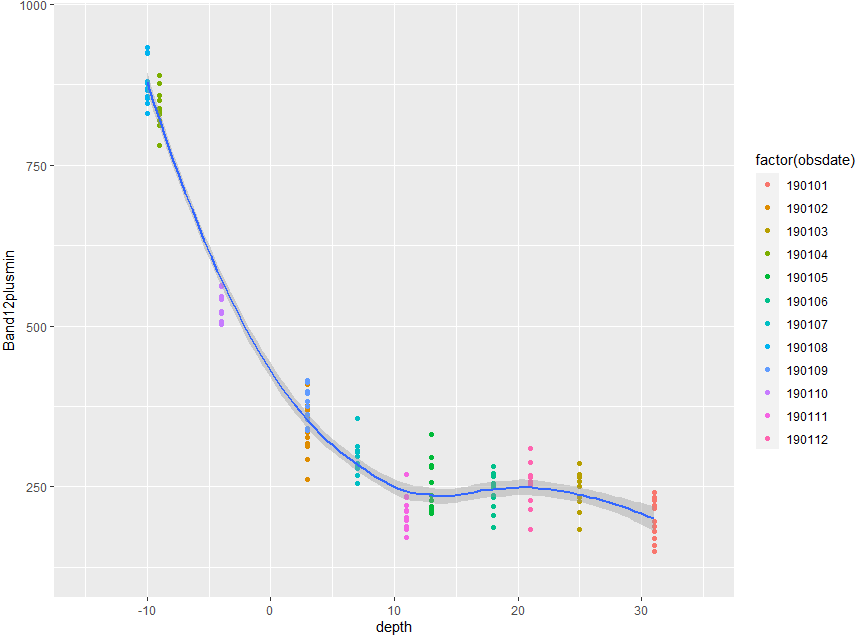
Here following structure, the first 25 lines and a scatterplot with a loess-line based on ALL observations
> str(df_Bandvals)
'data.frame': 144 obs. of 5 variables:
$ obsdate : int 190101 190101 190101 190101 190101 190101 190101 190101 190101 190101 ...
$ transsect : chr "A" "A" "A" "A" ...
$ PointNr : num 1 2 3 4 5 6 1 2 3 4 ...
$ depth : num 31 31 31 31 31 31 31 31 31 31 ...
$ Band12plusmin: num 169 241 229 159 221 196 188 216 233 149 ...
> df_Bandvals
obsdate transsect PointNr depth Band12plusmin
1 190101 A 1 31 169
2 190101 A 2 31 241
3 190101 A 3 31 229
4 190101 A 4 31 159
5 190101 A 5 31 221
6 190101 A 6 31 196
7 190101 B 1 31 188
8 190101 B 2 31 216
9 190101 B 3 31 233
10 190101 B 4 31 149
11 190101 B 5 31 169
12 190101 B 6 31 181
13 190102 A 1 3 356
14 190102 A 2 3 368
15 190102 A 3 3 293
16 190102 A 4 3 261
17 190102 A 5 3 313
18 190102 A 6 3 374
19 190102 B 1 3 327
20 190102 B 2 3 409
21 190102 B 3 3 369
22 190102 B 4 3 334
23 190102 B 5 3 376
24 190102 B 6 3 318
25 190103 A 1 25 183
The plot shows depth vs. Band12plusmin with an according loess-line. Point colors are assigned to the respective observation date (obsdate). Each observation date includes 12 observations.
Now, my basic question was: How to get a loess line based only on the lower 50%-quantile Band12plusmin-values of each observation date? Or in other words with referring to the plot: ggplot should only use the 6 lower points of each color for calculating the line.
And as mentioned before I would like to do the filtering or subsetting directly in ggplot without creating a new data object or subdataframe in advance.
I tried around with subsetting, but my problem in this case is that I cannot just specify a universal Band12plusmin-threshold as, of course, the 50%-treshold individually differs for each obsdate-group. I am quite new to R and ggplot, so, for now I failed to find a solution for that say class-individual-derived-threshold-conditionned filtering. May anybody help here?
Here the code of the dataframe and plot
obsdate<-rep(c(190101:190112),each=12, mode=factor)
transsect<-rep(rep(c("A","B"), each=6), 12)
PointNr<-rep(c(1,2,3,4,5,6), times=24)
depth<-rep(c(31,3,25,-9,13,18,7,-10,3,-4,11,21),each=12)
Band12<-rep(c(199,349,225,844,257,231,301,875,378,521,210,246), each=12)
set.seed(13423)
plusminRandom<-round(rnorm(144, mean=0, sd=33))
plusminRandom
Band12plusmin<-Band12 plusminRandom
df_Bandvals<-data.frame(obsdate, transsect, PointNr, depth, Band12plusmin)
str(df_Bandvals)
head(df_Bandvals, 20)
library (ggplot2)
ggplot(data=df_Bandvals, aes(x=depth, y=Band12plusmin))
scale_x_continuous(limits = c(-15, 35))
scale_y_continuous(limits = c(120, 960))
geom_point(aes(color=factor(obsdate)), size=1.5)
geom_smooth(method="loess")
CodePudding user response:
You should be able to use the data argument within geom_smooth()
ggplot(data = df_Bandvals, aes(x = depth, y = Band12plusmin))
scale_x_continuous(limits = c(-15, 35))
scale_y_continuous(limits = c(120, 960))
geom_point(aes(color = factor(obsdate)), size = 1.5)
geom_smooth(
data = df_Bandvals %>%
group_by(obsdate) %>%
filter(Band12plusmin < median(Band12plusmin)),
method = "loess"
)