I'm building a cluster with genetic algorithm optimization, but I'm having problems creating a legend for the cluster plot. This is my code. Please help me.
c1 = numpy.array([X[:, 0], X[:, 1]]).T
c2 = numpy.array([G[:, 0], G[:, 1]]).T
c3 = numpy.array([H[:, 0], H[:, 1]]).T
data = numpy.concatenate((c1, c2, c3), axis=0)
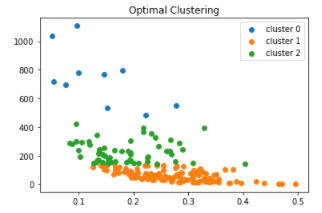
I have labeled this plot
matplotlib.pyplot.scatter(X[:, 0], X[:, 1], label="cluster 0")
matplotlib.pyplot.scatter(G[:, 0], G[:, 1], label="cluster 1")
matplotlib.pyplot.scatter(H[:, 0], H[:, 1], label="cluster 2")
matplotlib.pyplot.title("Optimal Clustering")
plt.legend()
matplotlib.pyplot.show()
def cluster_data(solution, solution_idx):
global num_clusters, feature_vector_length, data
cluster_centers = []
all_clusters_dists = []
clusters = []
clusters_sum_dist = []
for clust_idx in range(num_clusters):
cluster_centers.append(solution[feature_vector_length*clust_idx:feature_vector_length*(clust_idx 1)])
cluster_center_dists = euclidean_distance(data, cluster_centers[clust_idx])
all_clusters_dists.append(numpy.array(cluster_center_dists))
cluster_centers = numpy.array(cluster_centers)
all_clusters_dists = numpy.array(all_clusters_dists)
cluster_indices = numpy.argmin(all_clusters_dists, axis=0)
for clust_idx in range(num_clusters):
clusters.append(numpy.where(cluster_indices == clust_idx)[0])
if len(clusters[clust_idx]) == 0:
clusters_sum_dist.append(0)
else:
clusters_sum_dist.append(numpy.sum(all_clusters_dists[clust_idx, clusters[clust_idx]]))
clusters_sum_dist = numpy.array(clusters_sum_dist)
return cluster_centers, all_clusters_dists, cluster_indices, clusters, clusters_sum_dist
num_clusters = 3
feature_vector_length = data.shape[1]
num_genes = num_clusters * feature_vector_length
ga_instance = pygad.GA(num_generations=1000,
sol_per_pop=10,
init_range_low=0,
init_range_high=20,
num_parents_mating=5,
keep_parents=2,
num_genes=num_genes,
fitness_func=fitness_func,
suppress_warnings=True)
ga_instance.run()
best_solution, best_solution_fitness, best_solution_idx = ga_instance.best_solution()
print("Best solution is {bs}".format(bs=best_solution))
print("Fitness of the best solution is {bsf}".format(bsf=best_solution_fitness))
print("Best solution found after {gen} generations".format(gen=ga_instance.best_solution_generation))
cluster_centers, all_clusters_dists, cluster_indices, clusters, clusters_sum_dist = cluster_data(best_solution, best_solution_idx)
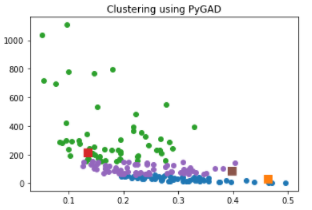
For this plot, can it be like the plot legend display that has been labeled from the beginning as above?
for cluster_idx in range(num_clusters):
cluster_x = data[clusters[cluster_idx], 0]
cluster_y = data[clusters[cluster_idx], 1]
matplotlib.pyplot.scatter(cluster_x, cluster_y)
matplotlib.pyplot.scatter(cluster_centers[cluster_idx, 0], cluster_centers[cluster_idx, 1], marker="s", s=100)
matplotlib.pyplot.title("Clustering using PyGAD")
matplotlib.pyplot.show()
I'm confused because for this plot display, the cluster_x and cluster_y data are separate. How can I create a legend based on the cluster that is formed, and also the cluster center?
Thank you in advance.
CodePudding user response:
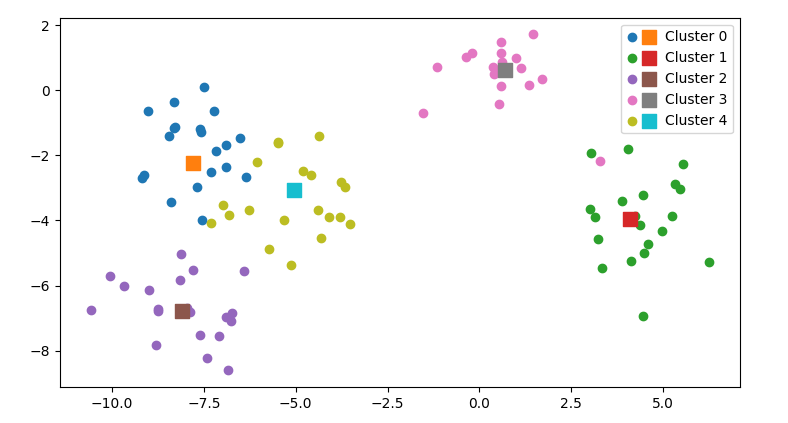
By adding label= to most matplotlib functions, a legend entry will be generated. The legend will be created by plt.legend().
Another possibility is to show the legend into two columns:
handles, labels = plt.gca().get_legend_handles_labels()
handles = [handles[i] for i in range(0, len(handles), 2)] [handles[i] for i in range(1, len(handles), 2)]
labels = [labels[i] for i in range(0, len(labels), 2)] [labels[i] for i in range(1, len(labels), 2)]
plt.legend(handles=handles, labels=labels, ncol=2)