Sincere apologies if my terminology is inaccurate, I am very new to R and programming in general (<1m experience). I was recently given the opportunity to do data analysis on a project I wish to write-up for a conference and could use some help.
I have a csv file (cida_ams_scc_csv) with patient data from a recent study. It's a dataframe, with columns of patient ID ('Cow ID'), location of sample ('QTR', either LH LF RH or RF), date ('Date', written DD/MM/YY), and the lab result from testing of the sample ('SCC', an integer).
For any given day, each of the four anatomic locations for each patient were sampled and tested. I want to find the average 'SCC' of the each of the locations for each of the patients, across all days the patient was sampled.
I was able to find the average SCC for each patient across all days and all anatomic sites using the code below.
aggregate(cida_ams_scc_csv$SCC, list(cida_ams_scc_csv$'Cow ID'), mean)
Now I want to add another "layer," where I see not just the patient's average, but the average of each patient for each of the 4 sample sites.
I honestly have no idea where to start. Please walk me through this in the simplest way possible, I will be eternally grateful.
CodePudding user response:
It is always better to provide a minimal reproducible example. But here the answer might be easy enough so its not necessary...
You can use the same code to do what you want. If we look at the aggregate documentation ?aggregate we find that the second argument by is
a list of grouping elements, each as long as the variables in the data frame x. The elements are coerced to factors before use.
Therefore running:
aggregate(mtcars$mpg, by = list(mtcars$cyl, mtcars$gear), mean)
Returns the "double grouped" means
In your case that means adding the "second layer" to the list you pass as value for the by parameter.
CodePudding user response:
I'd recommend dplyr for working with data frames - it's fast and friendly. Here's a way to calculate the average of each patient for each location using it:
# Load the package
library(dplyr)
# Make some fake data that looks similar to yours
cida_ams_scc_csv <- data.frame(
QTR=gl(
n = 4, k = 5, labels = c("LH", "LF", "RH", "RF"), length = 40
),
Date=rep(c("06/10/2021", "05/10/2021"), each=5),
SCC=runif(40),
Cow_ID=1:5)
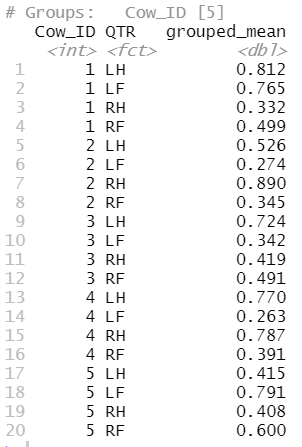
# Group by ID and QTR, then calculate the mean for each group
cida_ams_scc_csv %>%
group_by(Cow_ID, QTR) %>%
summarise(grouped_mean=mean(SCC))
which returns