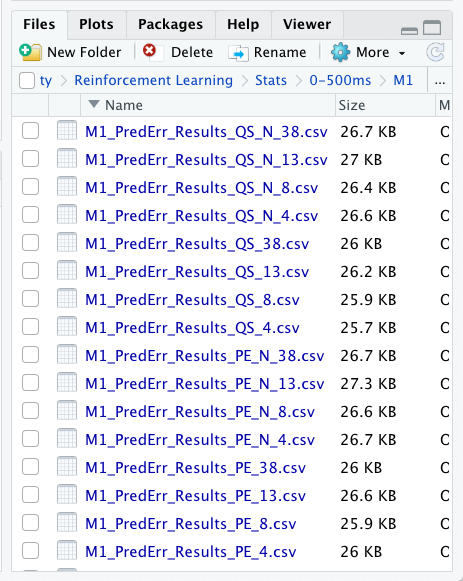
I am simply doing a for inside a for to read from different csv documents into a list of elements in R using the following code, were i take from two different variables, the frequency type freq <- c('4', '8', '13', '38','N_4','N_8','N_13','N_38') and the type of value typ <- c('PE','QS'). This two string variables are used for naming in the corresponding csv as seen in the working directory as seen in the image.
I am using the following code to read, from the same directory.
#Prepare data options
freq <- c('4', '8', '13', '38','N_4','N_8','N_13','N_38')
typ <- c('PE','QS')
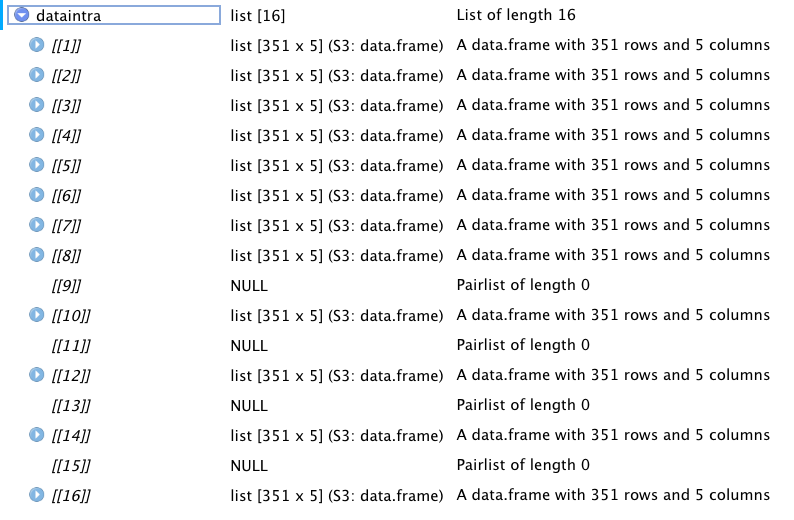
dataintra <- list()
for (j in 1:2){
for (k in 1:8){
dataintra[[j*k]] <- read.csv(sprintf('M1_PredErr_Results_%s_%s.csv', typ[j], freq[k]), sep=',', header=TRUE)
}
}
The problem is while I would be expecting a list of full list, I find different list numbers as NULL as can be seen in the following screenshot.
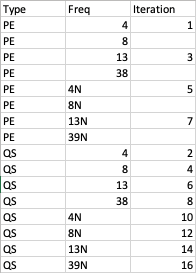
Trying to understand the logic behind the iteration, the for-for jumps in a weird way with odd numbers compared to even numbers, which makes the odds from 9 to 15 to be unable to complete. I am attaching the iteration order from every element of the list.
I would like to understand why R in a for-for has this erratic behaviour and what would be the correct way of doing this. I can think on other less efficient ways of doing this, for instance making an index vector with the two features attached iteration <- c('PE_4', 'PE_8', 'PE_13', 'PE_38','PE_N_4','PE_N_8','PE_N_13','PE_N_38','QS_4', 'QS_8', 'QS_13', 'QS_38','QS_N_4','QS_N_8','QS_N_13','QS_N_38') that makes the trick, but I would really like to understand the logic if anyone can help.
Thanks in advance!
CodePudding user response:
#Prepare data options
freq <- c('4', '8', '13', '38','N_4','N_8','N_13','N_38')
typ <- c('PE','QS')
I'd suggest using a named list (we can use the file names as identification)
dataintra <- list()
for (this_freq in freq){
for (this_typ in typ){
this_name <- sprintf('M1_PredErr_Results_%s_%s.csv', this_typ, this_freq)
dataintra[[this_name]] <- read.csv(this_name, sep=',', header=TRUE)
}
}
Or if you prefer a numeric index:
index <- 1
for (this_freq in freq){
for (this_typ in typ){
this_name <- sprintf('M1_PredErr_Results_%s_%s.csv', this_typ, this_freq)
dataintra[[index]] <- read.csv(this_name, sep=',', header=TRUE)
index <- index 1
}
}
CodePudding user response:
Your Problem is in the indexing of the List elements:
dataintra[[j*k]]
As for example 9 can only be made out of 3x3, the index 9 of the list will never be filled. For you only multiply (1:2)x(1:8)
You also overwrite multiple indices, for example index 8 is (1,8) and (4,2).
I think it would be easier with some functions R already provides, instead of using for-loops:
#Prepare data options
freq <- c('4', '8', '13', '38','N_4','N_8','N_13','N_38')
typ <- c('PE','QS')
# get all combinations of typ and freq
input<- expand.grid(typ,freq)
## get a List of all the adresses
# mapply iterates over two vectors (Var1 and Var2 of the input dataframe)
# and performs a function on them
csv_adresses <- mapply(function(x,y){sprintf('M1_PredErr_Results_%s_%s.csv',x,y)},
input$Var1,input$Var2)
# read all the csv files of the adresses
dataintra <- sapply(csv_adresses,read.csv,header=TRUE)