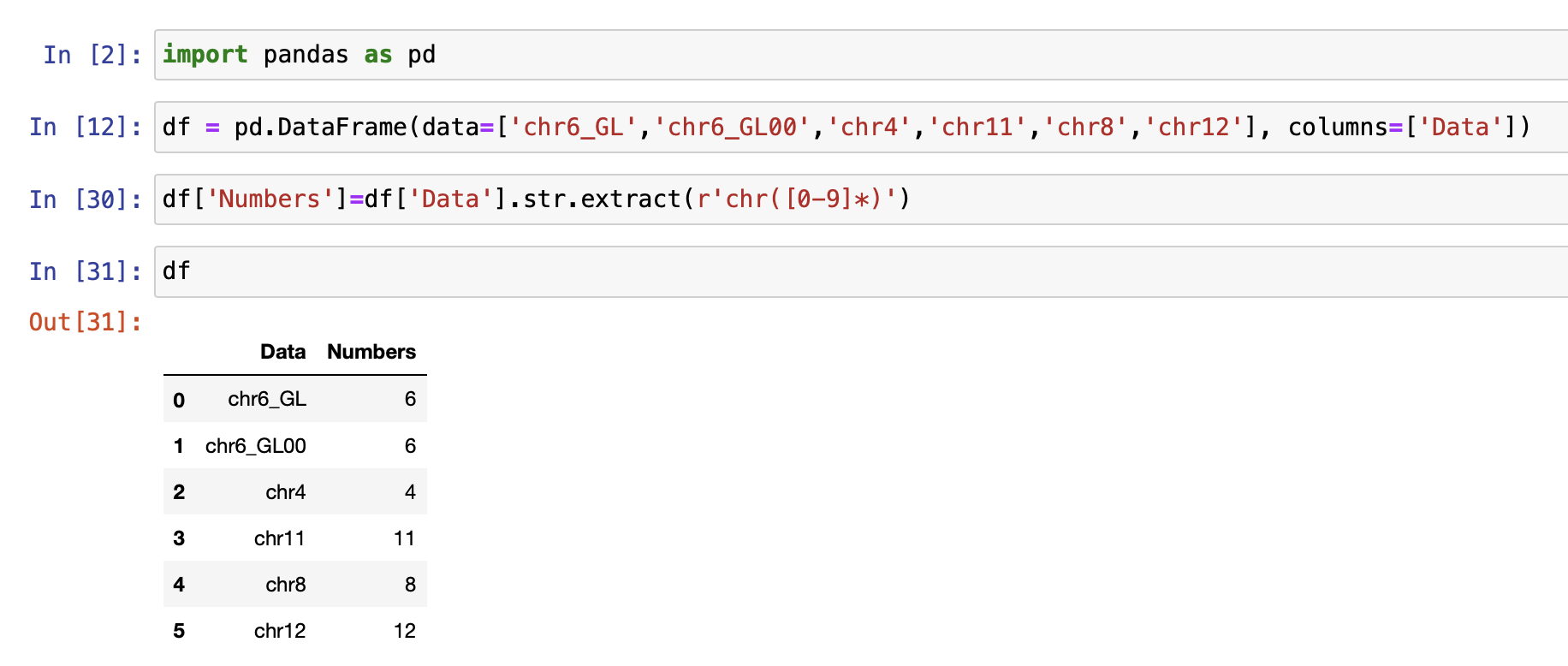
I have a data frame that has a column containing the chromosome details (1 to 22). I would like to create another column with only Chr numbers
CodePudding user response:
Please find below a solution with the package data.table:
REPREX
- Code
library(data.table)
library(stringr)
DT[, Chr_ID := lapply(.SD, str_extract,"(?<=^chr)\\d "), .SDcols = "chromosome"]
- Output
DT
#> chromosome Chr_ID
#> 1: chr6_GL000253v2_alt 6
#> 2: chr6_GL000254v2_alt 6
#> 3: chr6_GL000255v2_alt 6
#> 4: chr6_GL000256v2_alt 6
#> 5: chr4 4
#> 6: chr11 11
#> 7: chr8 8
#> 8: chr12 12
#> 9: chr2 2
#> 10: chr12 12
#> 11: chr4 4
#> 12: chr6 6
#> 13: chr15 15
#> 14: chr4 4
#> 15: chr2 2
- Your data
DT <- data.table(chromosome = c("chr6_GL000253v2_alt", "chr6_GL000254v2_alt",
"chr6_GL000255v2_alt", "chr6_GL000256v2_alt", "chr4", "chr11",
"chr8", "chr12", "chr2", "chr12", "chr4", "chr6", "chr15", "chr4",
"chr2"))
DT
#> chromosome
#> 1: chr6_GL000253v2_alt
#> 2: chr6_GL000254v2_alt
#> 3: chr6_GL000255v2_alt
#> 4: chr6_GL000256v2_alt
#> 5: chr4
#> 6: chr11
#> 7: chr8
#> 8: chr12
#> 9: chr2
#> 10: chr12
#> 11: chr4
#> 12: chr6
#> 13: chr15
#> 14: chr4
#> 15: chr2