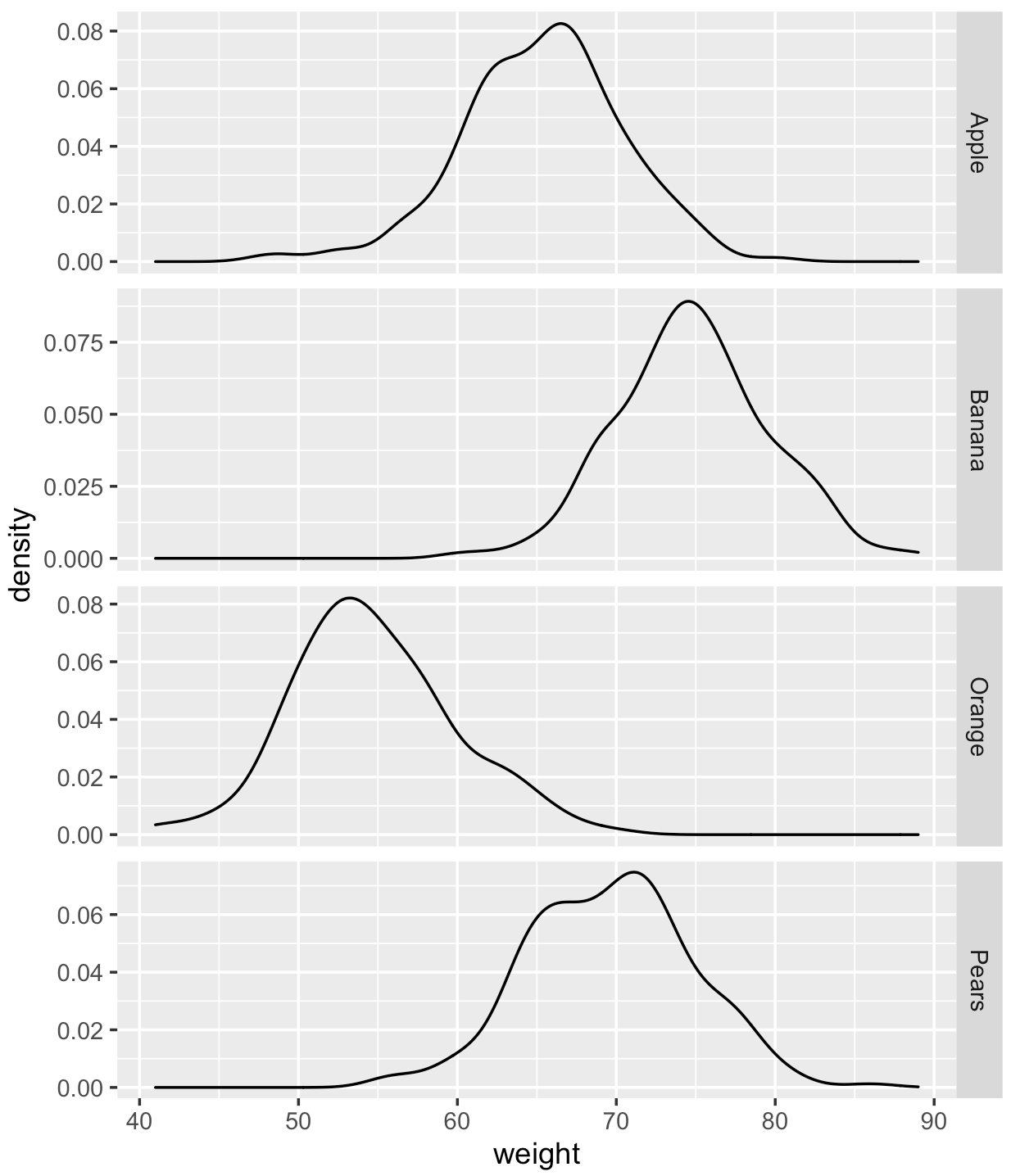
How can I extract the values matrix of each density plot?
For example, I am interested in that when weight = 71, what will the density of orange, apple, pears, banana be ?
Below is the minimal example:
library(ggplot2)
set.seed(1234)
df = data.frame(
fruits = factor(rep(c("Orange", "Apple", "Pears", "Banana"), each = 200)),
weight = round(c(rnorm(200, mean = 55, sd=5),
rnorm(200, mean=65, sd=5),
rnorm(200, mean=70, sd=5),
rnorm(200, mean=75, sd=5)))
)
dim(df) [1] 800 2
ggplot(df, aes(x = weight))
geom_density()
facet_grid(fruits ~ ., scales = "free", space = "free")
CodePudding user response:
I think the underlying density in ggplot2 is similar to stats::density, and thus we can use that to build the same info
df %>%
group_by(fruits) %>%
nest() %>%
ungroup() %>%
mutate(density = data %>% map(. %>%
`[[`("weight") %>%
density.default)) %>%
hoist(density, "x", "y") %>%
select(-density, -data) %>%
unnest(c(x,y)) %>%
group_by(fruits) %>%
slice_min(abs(x - 71), n = 1) %>%
ungroup() %>%
identity()
# A tibble: 5 x 3
fruits x y
<fct> <dbl> <dbl>
1 Apple 71.0 0.0409
2 Banana 71.0 0.0574
3 Orange 71.0 0.00131
4 Pears 71.0 0.0747
5 Pears 71.0 0.0747
If you don't think that's enough, then this is an extraction from the plot, that you specified:
gg_density_plot %>%
ggplot_build() %>%
`[[`("data") %>%
`[[`(1) %>%
as_tibble() %>%
# glimpse() %>%
# count(PANEL) # panel is fruit
group_by(PANEL) %>%
slice_min(abs(x - 71), n = 1) %>%
ungroup() %>%
select(PANEL, x, y, density)
# A tibble: 4 x 4
PANEL x y density
<fct> <dbl> <dbl> <dbl>
1 1 71.0 0.0410 0.0410
2 2 71.0 0.0568 0.0568
3 3 71.0 0.00135 0.00135
4 4 71.0 0.0747 0.0747
CodePudding user response:
Save the plot in a variable, build the data structure with ggplot_build and split the data by panel. Then interpolate with approx to get the new values.
g <- ggplot(df, aes(x = weight))
geom_density()
facet_grid(fruits ~ ., scales = "free", space = "free")
p <- ggplot_build(g)
# These are the columns of interest
p$data[[1]]$x
p$data[[1]]$density
p$data[[1]]$PANEL
Split the list member p$data[[1]] by panel but keep only the x and density values. Then loop through the split data to interpolate by group of fruit.
sp <- split(p$data[[1]][c("x", "density")], p$data[[1]]$PANEL)
new_weight <- 71
sapply(sp, \(DF){
with(DF, approx(x, density, xout = new_weight))
})
# 1 2 3 4
#x 71 71 71 71
#y 0.04066888 0.05716947 0.001319164 0.07467761
Or, without splitting the data previously, use by.
b <- by(p$data[[1]][c("x", "density")], p$data[[1]]$PANEL, \(DF){
with(DF, approx(x, density, xout = new_weight))
})
do.call(rbind, lapply(b, as.data.frame))
# x y
#1 71 0.040668880
#2 71 0.057169474
#3 71 0.001319164
#4 71 0.074677607