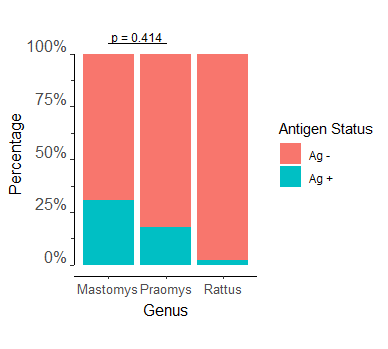
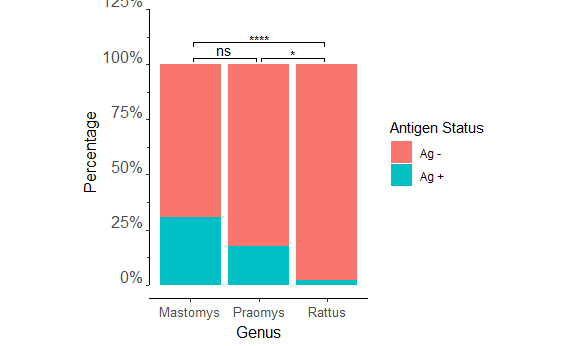
I am trying to add multiple p-values to my stacked bar chart. I can get a single p-value added to my data with the following code:
library(tidyverse)
library(ggprism)
library(rstatix)
RDT_Pos_genus_pval <- table(RDT_genus_ratio$Ag_RDT, RDT_genus_ratio$Genus_Cytb) %>% ##filtered to one group for example purposes
fisher.test() %>%
.$p.value %>%
signif(3)
Pos_genus_pval <- data.frame(
group1 = "Mastomys",
group2 = "Praomys",
label = RDT_Pos_genus_pval,
y.position = 1.05)
RDT_genus_ratio_bar <- ggplot(RDT_genus_ratio)
geom_bar(mapping = aes(x = Genus_Cytb, fill = Ag_RDT), position = "fill")
labs(y = "Percentage", x = "Genus")
scale_y_continuous(labels = scales::percent, guide = "prism_offset_minor")
scale_x_discrete()
scale_fill_discrete(name = "Antigen Status", labels = c("FALSE" = "Ag -", "TRUE" = "Ag "))
theme_classic()
theme(aspect.ratio = 2/1.5, plot.title = element_text(hjust = 0.5, size = 16),
axis.text.y = element_text(size = 12, vjust = -0.2), axis.text.x = element_text(size = 10),
axis.title = element_text(size = 12), axis.title.x.bottom = element_text(size = 12, vjust = 0.5))
add_pvalue(Pos_genus_pval, label = "p = {label}")
However, I'd like to add pairwise fisher tests to all three groups. I know with the rstatix package I should be able to do this by creating a dataframe specifically for this, then adding it in place of Pos_genus_pval in the add_pvalue line. However, I keep getting stuck on the add_y_position part (see below)
asdf <- table(RDT_genus_ratio$Ag_RDT, RDT_genus_ratio$Genus_Cytb) %>%
pairwise_fisher_test()
# A tibble: 3 x 6
group1 group2 n p p.adj p.adj.signif
* <chr> <chr> <int> <dbl> <dbl> <chr>
1 Mastomys Praomys 316 4.14e- 1 4.14e- 1 ns
2 Mastomys Rattus 491 7.44e-18 2.23e-17 ****
3 Praomys Rattus 209 1.29e- 2 2.58e- 2 *
asdf <- table(RDT_genus_ratio$Ag_RDT, RDT_genus_ratio$Genus_Cytb) %>%
pairwise_fisher_test() %>%
add_y_position(formula = group1 ~ group2) ##this line is where I'm getting stuck
Error in add_y_position(., formula = group1 ~ group2) :
data and formula arguments should be specified.