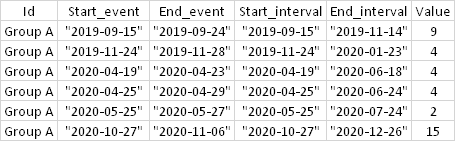
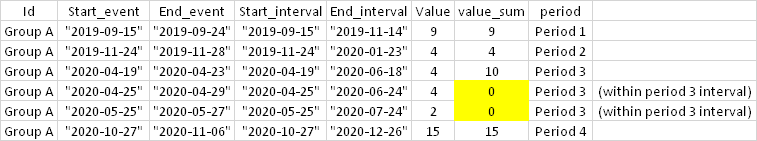
I have the following dataset:
Data:
I would like to sum, by group, the values of all events that started within 60 days (already calculated interval = Start_interval, End_interval) without adding the same row in more than one interval.
Expected output:
I did research and found some solutions that allowed me to obtain, until then, the results displayed below, very close to what I expected. For example: 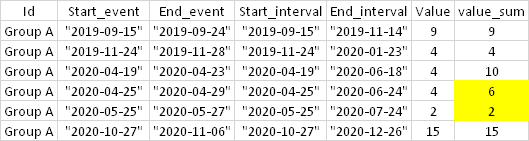
Does anyone have any suggestions?
Reproducible example:
# Input data
data <- data.table(id = c("Group A", "Group A", "Group A", "Group A",
"Group A", "Group A"),
start_date_event = c("2019-09-15",
"2019-11-24",
"2020-04-19",
"2020-04-25",
"2020-05-25",
"2020-10-27"),
end_date_event = c("2019-09-24",
"2019-11-28",
"2020-04-23",
"2020-04-29",
"2020-05-27",
"2020-11-06"),
start_interval = c("2019-09-15",
"2019-11-24",
"2020-04-19",
"2020-04-25",
"2020-05-25",
"2020-10-27"),
end_interval = c("2019-11-14",
"2020-01-23",
"2020-06-18",
"2020-06-24",
"2020-07-24",
"2020-12-26"),
value = c(9, 4, 4, 4, 2, 15))
# Convert to date
data <- data %>%
dplyr::mutate(start_date_event = as.Date(start_date_event)) %>%
dplyr::mutate(end_date_event = as.Date(end_date_event)) %>%
dplyr::mutate(start_interval = as.Date(start_interval)) %>%
dplyr::mutate(end_interval = as.Date(end_interval))
# Calculating with non-equi join
temp <- data[data,
on = .(start_date_event <= end_interval,
end_date_event >= start_interval)][,
.(value_sum = sum(value)),
by = .(id, start_date_event)]
# Get all
data <- merge(data, temp, all.x = T,
by.x = c("id", "end_interval"),
by.y = c("id", "start_date_event"))
Thanks!
CodePudding user response:
Here's a seemingly-complicated approach that gets your results:
data[, rn := seq_len(.N)
][data, on = .(id, start_date_event >= start_interval, end_date_event <= end_interval)
][, z := fifelse(rleid(i.rn) > 1, 0, value), by = rn
][, value_sum := sum(z), by = i.rn
][, .SD[1,], .SDcols = patterns("^.[^.]"), by=.(i.rn)
][, c("rn", "i.rn") := NULL ]
# id start_date_event end_date_event start_interval end_interval value value_sum
# <char> <Date> <Date> <Date> <Date> <num> <num>
# 1: Group A 2019-09-15 2019-11-14 2019-09-15 2019-11-14 9 9
# 2: Group A 2019-11-24 2020-01-23 2019-11-24 2020-01-23 4 4
# 3: Group A 2020-04-19 2020-06-18 2020-04-19 2020-06-18 4 10
# 4: Group A 2020-04-25 2020-06-24 2020-04-25 2020-06-24 4 0
# 5: Group A 2020-05-25 2020-07-24 2020-05-25 2020-07-24 2 0
# 6: Group A 2020-10-27 2020-12-26 2020-10-27 2020-12-26 15 15
CodePudding user response:
Here is one option..
First, change to dates (this is like your dplyr/mutate statement above)
data <- cbind(data[, .(id, value)], data[, lapply(.SD, as.Date), .SDcols = c(2,3,4,5)])
Add an event_id column, within group
data[order(id,start_date_event), event_id:=1:.N, id]
Get a table of unique "periods" by id and key the table, for use in foverlaps
periods <- data[, .(id, start_interval, end_interval)][, period:=1:.N, by=id]
setkey(periods, id, start_interval, end_interval)
Use fast overlaps to associate a period with each event, then get the minimum period for each event, and the sum of values by each period
period_id <- foverlaps(data, periods, by.x = c("id", "start_date_event", "end_date_event"))
Create the value sum column via these steps
# Get the value_sums, by merging the minimum period by event
# with the sum over the values by period
value_sums = period_id[,.(period = min(period)),
by=.(id, event_id)][
period_id[
, .(value_sum = sum(value)),
by = .(id, period)],
on=.(id, period), nomatch=0]
# convert the value sum column to zero if it is not the first row, by associated period
value_sums[order(id,event_id, period),value_sum:=value_sum*((1:.N)==1), by=.(id, period)]
# merge back on to data (dropping the period column)
data[value_sums[, !c("period")], on=.(id,event_id)]
Output:
id value start_date_event end_date_event start_interval end_interval event_id value_sum
1: Group A 9 2019-09-15 2019-09-24 2019-09-15 2019-11-14 1 9
2: Group A 4 2019-11-24 2019-11-28 2019-11-24 2020-01-23 2 4
3: Group A 4 2020-04-19 2020-04-23 2020-04-19 2020-06-18 3 10
4: Group A 4 2020-04-25 2020-04-29 2020-04-25 2020-06-24 4 0
5: Group A 2 2020-05-25 2020-05-27 2020-05-25 2020-07-24 5 0
6: Group A 15 2020-10-27 2020-11-06 2020-10-27 2020-12-26 6 15