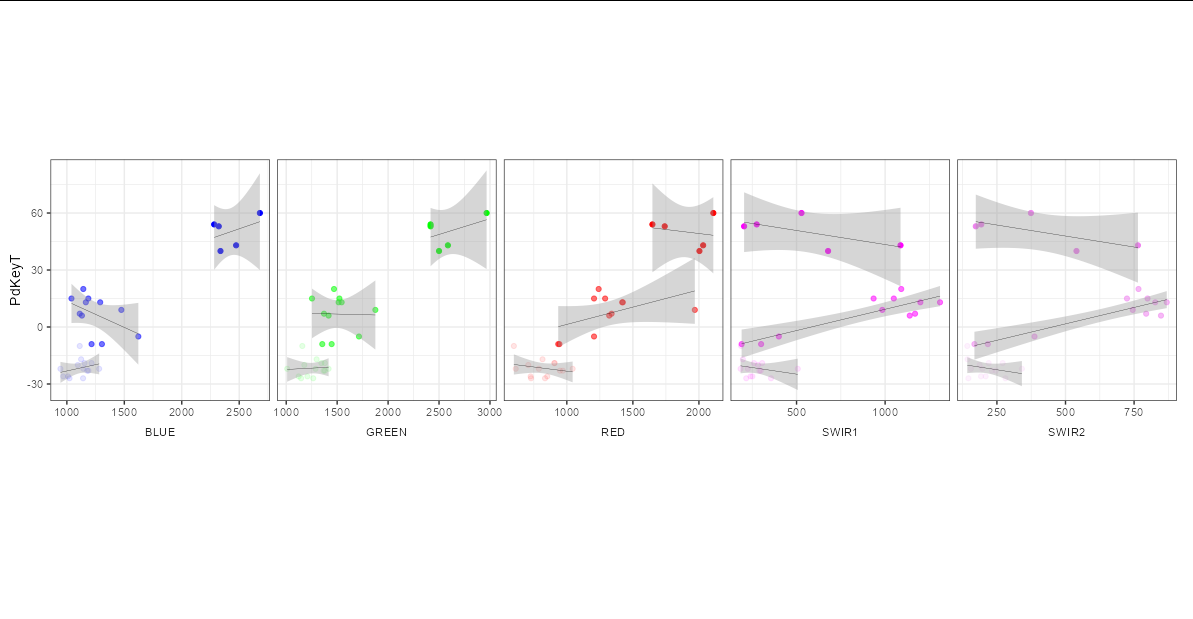
My dataframe loopsubset_created contains 30 observations of 45 variables. (Below you will find str(loopsubset_created) and a dput(loopsubset_created) sample).
Now I would like to create scatter plots of the PdKeyT-Variable (y) versus five of the band-value-variables (BLUE, GREEN, RED, SWIR1, SWIR2) (x) with
- each variable in ONE panel
- all panels aligned in ONE row
- using the
PdKeyTvariable as common y-axis.
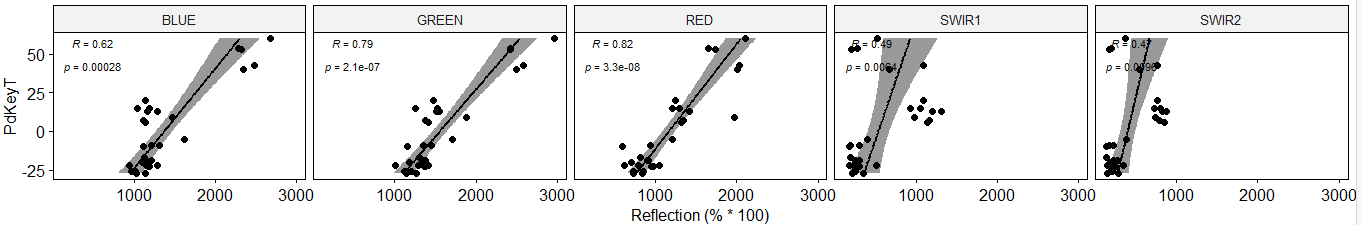
In the end it should basically look like this:
(I did this with ggscatter, but for flexibility reasons I would prefer basically using ggplot)
Here now my issue:
When trying with ggplot, I do not find the right way for the above showed arrangement, as I cannot figure out the right code for separating/grouping by variables. I found hundreds of tutorials for facetting by multiple categorial values within a variable, but not by multiple variables.
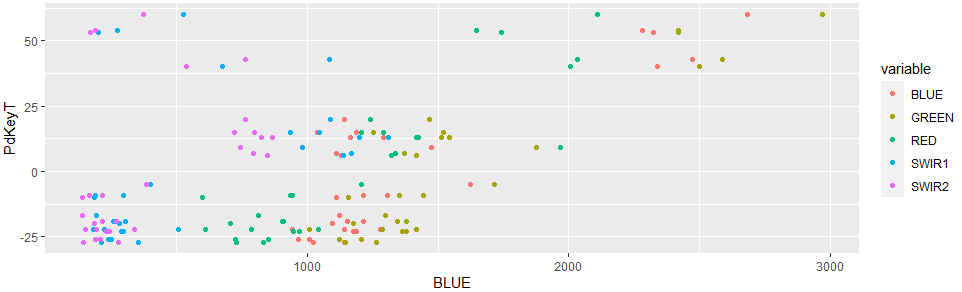
With the following code
ggplot(loopsubset_created, aes(y = PdKeyT))
geom_point(aes(x = BLUE, col = "BLUE"))
geom_point(aes(x = GREEN, col = "GREEN"))
geom_point(aes(x = RED, col = "RED"))
geom_point(aes(x = SWIR1, col = "SWIR1"))
geom_point(aes(x = SWIR2, col = "SWIR2"))
I came to this basic result
Here the basic question:
Now, I would like to arrange the 5 layers seperately in one row according to the above depicted way
Anybody an idea for me?
Plus some information around the question:
Though the following aspects are not directly part of my question, I'd like to describe my final idea of the plot (in order to avoid that your advices may clash with further requirements):
Each panel should include
- Spearman corr value and according p-value (as shown above) and
- additionaly Pearson corr value and according p-value
- Linear regression with conf. interval (as shown above) or other type of regression line (not shown)
- Points should be couloured by variable (BLUE=bLue, RED= red; GREEN=green, SWIR1 2 by some other coulours, e.g. magenta and violet)
- later on points and regressionlines should be subdived by ranges of
PdKeyT(e.g. below -10, -10-to 30, and above 30) with using differnt brightness values of variable basic colours (blue, green, ...), analogouos to this:
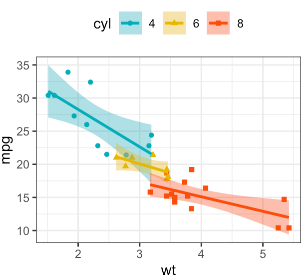
- All panels should use ONE common y-axis at the left as explained
- And I would like to adpat the x-axes by the range of the respecitve variable (e.g. range for BLUE, GREEN and RED from 500 to 3000 and the SWIRs from 0 to 1500
Finally some information and sample of my data
> str(loopsubset_created)
'data.frame': 30 obs. of 45 variables:
$ Site_ID : chr "A" "A" "A" "A" ...
$ Spot_Nr : chr "1" "1" "1" "1" ...
$ Transkt_Nr : chr "2" "2" "2" "2" ...
$ Point_Nr : chr "4" "4" "4" "4" ...
$ n : int 30 30 30 30 30 30 30 30 30 30 ...
$ rank : int 3 3 3 3 3 3 3 3 3 3 ...
$ Tile : chr "1008" "1008" "1008" "1008" ...
$ Date : int 20190208 20190213 20190215 20190218 20190223 20190228 20190302 20190305 20190315 20190320 ...
$ id : chr "22" "22" "22" "22" ...
$ Point_ID : chr "1022" "1022" "1022" "1022" ...
$ Site_Nr : chr "1" "1" "1" "1" ...
$ Point_x : num 356251 356251 356251 356251 356251 ...
$ Point_y : num 5132881 5132881 5132881 5132881 5132881 ...
$ Classification : num 7 7 7 7 7 7 7 7 7 7 ...
$ Class_Derived : chr "WW" "WW" "WW" "WW" ...
$ BLUE : num 1112 1095 944 1144 1141 ...
$ GREEN : num 1158 1178 1009 1288 1265 ...
$ RED : num 599 708 613 788 835 ...
$ REDEDGE1 : num 359 520 433 576 665 761 618 598 881 619 ...
$ REDEDGE2 : num 83 82 65 169 247 404 116 118 532 162 ...
$ REDEDGE3 : num 73 116 81 142 233 391 56 171 538 131 ...
$ BROADNIR : num 44 93 60 123 262 349 74 113 560 125 ...
$ NIR : num 37 70 66 135 215 313 110 135 504 78 ...
$ SWIR1 : num 187 282 184 225 356 251 240 216 507 197 ...
$ SWIR2 : num 142 187 155 197 281 209 192 146 341 143 ...
$ Quality.assurance.information: num 26664 10272 10272 10272 8224 ...
$ Q00_VAL : num 0 0 0 0 0 0 0 0 0 0 ...
$ Q01_CS1 : num 0 0 0 0 0 0 0 0 0 0 ...
$ Q02_CSS : num 0 0 0 0 0 0 0 0 0 0 ...
$ Q03_CSH : num 1 0 0 0 0 0 0 0 1 0 ...
$ Q04_SNO : num 0 0 0 0 0 0 0 0 0 0 ...
$ Q05_WAT : num 1 1 1 1 1 1 1 1 1 1 ...
$ Q06_AR1 : num 0 0 0 0 0 0 0 0 0 0 ...
$ Q07_AR2 : num 0 0 0 0 0 0 0 0 0 0 ...
$ Q08_SBZ : num 0 0 0 0 0 0 0 0 0 0 ...
$ Q09_SAT : num 0 0 0 0 0 0 0 0 0 0 ...
$ Q10_ZEN : num 0 0 0 0 0 0 0 0 0 0 ...
$ Q11_IL1 : num 1 1 1 1 0 0 0 0 0 0 ...
$ Q12_IL2 : num 0 0 0 0 0 0 0 0 0 0 ...
$ Q13_SLO : num 1 1 1 1 1 1 1 1 1 1 ...
$ Q14_VAP : num 1 0 0 0 0 0 0 0 1 0 ...
$ Q15_WDC : num 0 0 0 0 0 0 0 0 0 0 ...
$ PdMax : int -7 -19 -20 -22 -24 -25 -26 -25 -21 -15 ...
$ PdMin : int -13 -23 -24 -26 -28 -29 -29 -28 -24 -20 ...
$ PdKeyT : int -10 -20 -22 -22 -27 -26 -26 -27 -22 -17 ...
loopsubset_created <- structure(list(Site_ID = c("A", "A", "A", "A", "A", "A", "A",
"A", "A", "A", "A", "A", "A", "A", "A", "A", "A", "A", "A", "A",
"A", "A", "A", "A", "A", "A", "A", "A", "A", "A"), Spot_Nr = c("1",
"1", "1", "1", "1", "1", "1", "1", "1", "1", "1", "1", "1", "1",
"1", "1", "1", "1", "1", "1", "1", "1", "1", "1", "1", "1", "1",
"1", "1", "1"), Transkt_Nr = c("2", "2", "2", "2", "2", "2",
"2", "2", "2", "2", "2", "2", "2", "2", "2", "2", "2", "2", "2",
"2", "2", "2", "2", "2", "2", "2", "2", "2", "2", "2"), Point_Nr = c("4",
"4", "4", "4", "4", "4", "4", "4", "4", "4", "4", "4", "4", "4",
"4", "4", "4", "4", "4", "4", "4", "4", "4", "4", "4", "4", "4",
"4", "4", "4"), n = c(30L, 30L, 30L, 30L, 30L, 30L, 30L, 30L,
30L, 30L, 30L, 30L, 30L, 30L, 30L, 30L, 30L, 30L, 30L, 30L, 30L,
30L, 30L, 30L, 30L, 30L, 30L, 30L, 30L, 30L), rank = c(3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L), Tile = c("1008",
"1008", "1008", "1008", "1008", "1008", "1008", "1008", "1008",
"1008", "1008", "1008", "1008", "1008", "1008", "1008", "1008",
"1008", "1008", "1008", "1008", "1008", "1008", "1008", "1008",
"1008", "1008", "1008", "1008", "1008"), Date = c(20190208L,
20190213L, 20190215L, 20190218L, 20190223L, 20190228L, 20190302L,
20190305L, 20190315L, 20190320L, 20190322L, 20190325L, 20190330L,
20190401L, 20190416L, 20190419L, 20190421L, 20190501L, 20190506L,
20190524L, 20190531L, 20190603L, 20190620L, 20190625L, 20190630L,
20190705L, 20190710L, 20190809L, 20190814L, 20190903L), id = c("22",
"22", "22", "22", "22", "22", "22", "22", "22", "22", "22", "22",
"22", "22", "22", "22", "22", "22", "22", "22", "22", "22", "22",
"22", "22", "22", "22", "22", "22", "22"), Point_ID = c("1022",
"1022", "1022", "1022", "1022", "1022", "1022", "1022", "1022",
"1022", "1022", "1022", "1022", "1022", "1022", "1022", "1022",
"1022", "1022", "1022", "1022", "1022", "1022", "1022", "1022",
"1022", "1022", "1022", "1022", "1022"), Site_Nr = c("1", "1",
"1", "1", "1", "1", "1", "1", "1", "1", "1", "1", "1", "1", "1",
"1", "1", "1", "1", "1", "1", "1", "1", "1", "1", "1", "1", "1",
"1", "1"), Point_x = c(356250.781, 356250.781, 356250.781, 356250.781,
356250.781, 356250.781, 356250.781, 356250.781, 356250.781, 356250.781,
356250.781, 356250.781, 356250.781, 356250.781, 356250.781, 356250.781,
356250.781, 356250.781, 356250.781, 356250.781, 356250.781, 356250.781,
356250.781, 356250.781, 356250.781, 356250.781, 356250.781, 356250.781,
356250.781, 356250.781), Point_y = c(5132880.701, 5132880.701,
5132880.701, 5132880.701, 5132880.701, 5132880.701, 5132880.701,
5132880.701, 5132880.701, 5132880.701, 5132880.701, 5132880.701,
5132880.701, 5132880.701, 5132880.701, 5132880.701, 5132880.701,
5132880.701, 5132880.701, 5132880.701, 5132880.701, 5132880.701,
5132880.701, 5132880.701, 5132880.701, 5132880.701, 5132880.701,
5132880.701, 5132880.701, 5132880.701), Classification = c(7,
7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7,
7, 7, 7, 7, 7, 7, 7, 7), Class_Derived = c("WW", "WW", "WW",
"WW", "WW", "WW", "WW", "WW", "WW", "WW", "WW", "WW", "WW", "WW",
"WW", "WW", "WW", "WW", "WW", "WW", "WW", "WW", "WW", "WW", "WW",
"WW", "WW", "WW", "WW", "WW"), BLUE = c(1112, 1095, 944, 1144,
1141, 1010, 968, 1023, 1281, 1124, 1215, 1154, 1188, 1177, 1622,
1305, 1215, 2282, 2322, 2337, 2680, 2473, 1143, 1187, 1165, 1040,
1290, 1112, 1474, 1131), GREEN = c(1158, 1178, 1009, 1288, 1265,
1208, 1122, 1146, 1416, 1298, 1379, 1345, 1379, 1366, 1714, 1446,
1354, 2417, 2417, 2500, 2967, 2587, 1469, 1522, 1544, 1253, 1514,
1371, 1875, 1416), RED = c(599, 708, 613, 788, 835, 852, 726,
729, 1044, 816, 905, 908, 948, 970, 1206, 944, 935, 1648, 1741,
2004, 2109, 2032, 1241, 1290, 1419, 1206, 1424, 1339, 1969, 1321
), REDEDGE1 = c(359, 520, 433, 576, 665, 761, 618, 598, 881,
619, 722, 771, 829, 823, 937, 725, 759, 1327, 1395, 1756, 1718,
1753, 1533, 1528, 1683, 1335, 1605, 1499, 2016, 1592), REDEDGE2 = c(83,
82, 65, 169, 247, 404, 116, 118, 532, 162, 183, 218, 285, 200,
514, 182, 230, 568, 531, 1170, 780, 1101, 1192, 1174, 1250, 949,
1121, 1127, 1382, 1159), REDEDGE3 = c(73, 116, 81, 142, 233,
391, 56, 171, 538, 131, 205, 137, 321, 253, 503, 193, 214, 564,
527, 1192, 698, 1177, 1203, 1259, 1341, 1049, 1146, 1216, 1416,
1188), BROADNIR = c(44, 93, 60, 123, 262, 349, 74, 113, 560,
125, 121, 211, 325, 221, 480, 184, 178, 461, 435, 1067, 570,
1023, 961, 966, 964, 844, 764, 993, 1197, 834), NIR = c(37, 70,
66, 135, 215, 313, 110, 135, 504, 78, 115, 216, 197, 163, 462,
113, 165, 392, 349, 1006, 574, 1092, 1153, 1143, 1128, 961, 1033,
1027, 1164, 1086), SWIR1 = c(187, 282, 184, 225, 356, 251, 240,
216, 507, 197, 306, 260, 298, 290, 400, 190, 300, 275, 204, 678,
528, 1087, 1091, 1049, 1310, 935, 1199, 1169, 984, 1139), SWIR2 = c(142,
187, 155, 197, 281, 209, 192, 146, 341, 143, 271, 220, 246, 232,
387, 168, 217, 193, 173, 540, 374, 764, 766, 799, 869, 724, 827,
794, 745, 848), Quality.assurance.information = c(26664, 10272,
10272, 10272, 8224, 8224, 8224, 8224, 24616, 8224, 8224, 8224,
32, 8224, 8288, 24616, 8224, 8240, 48, 8208, 8240, 8192, 8192,
24648, 8192, 8192, 8192, 8192, 0, 8224), Q00_VAL = c(0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0), Q01_CS1 = c(0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0),
Q02_CSS = c(0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0), Q03_CSH = c(1,
0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0,
0, 0, 0, 1, 0, 0, 0, 0, 0, 0), Q04_SNO = c(0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 1, 1, 0, 0, 0,
0, 0, 0, 0, 0, 0), Q05_WAT = c(1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 0, 1, 0, 0, 0, 0, 0, 0, 0,
0, 1), Q06_AR1 = c(0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0), Q07_AR2 = c(0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0), Q08_SBZ = c(0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0), Q09_SAT = c(0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0), Q10_ZEN = c(0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0), Q11_IL1 = c(1,
1, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0), Q12_IL2 = c(0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0), Q13_SLO = c(1, 1, 1, 1, 1, 1, 1, 1, 1,
1, 1, 1, 0, 1, 1, 1, 1, 1, 0, 1, 1, 1, 1, 1, 1, 1, 1, 1,
0, 1), Q14_VAP = c(1, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0,
0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0), Q15_WDC = c(0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 0, 0, 0, 0), PdMax = c(-7L, -19L, -20L,
-22L, -24L, -25L, -26L, -25L, -21L, -15L, -19L, -17L, -23L,
-22L, -4L, -7L, -8L, 55L, 57L, 47L, 67L, 44L, 21L, 18L, 13L,
16L, 16L, 9L, 12L, 11L), PdMin = c(-13L, -23L, -24L, -26L,
-28L, -29L, -29L, -28L, -24L, -20L, -22L, -22L, -26L, -26L,
-7L, -11L, -11L, 46L, 47L, 36L, 52L, 37L, 17L, 14L, 9L, 11L,
9L, 5L, 5L, 2L), PdKeyT = c(-10L, -20L, -22L, -22L, -27L,
-26L, -26L, -27L, -22L, -17L, -19L, -19L, -23L, -23L, -5L,
-9L, -9L, 54L, 53L, 40L, 60L, 43L, 20L, 15L, 13L, 15L, 13L,
7L, 9L, 6L)), row.names = 198:227, class = "data.frame")
CodePudding user response:
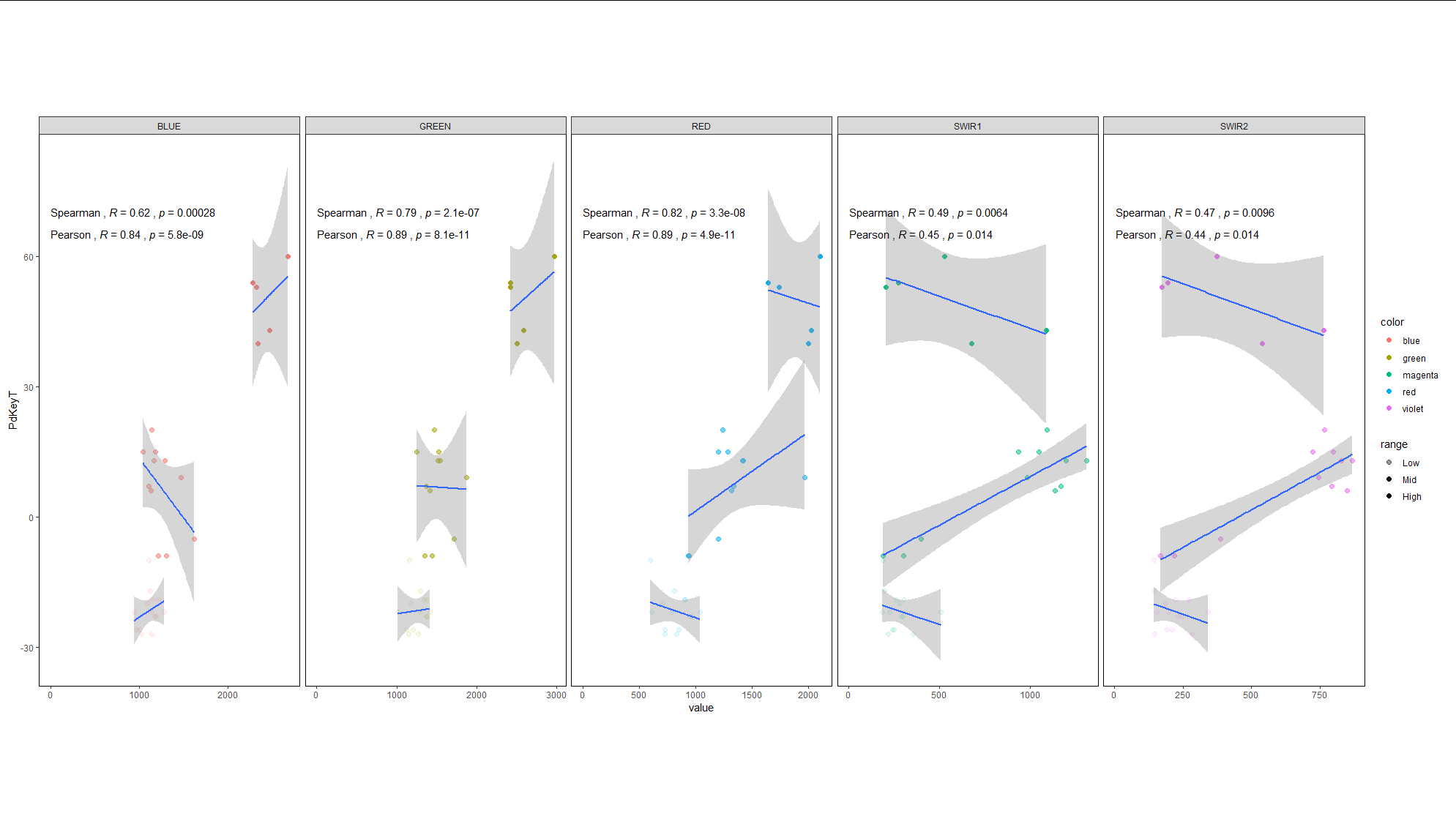
Update:
To fulfill your last task me could make use of the code that is from Allan Cameron: adding another column to set the cuts mutate(range = cut(PdKeyT, c(-Inf, -10, 30, Inf), c("Low", "Mid", "High"))) %>% (this code was provided by Allan Cameron)
library(tidyverse)
library(ggpubr)
df_long_list <- loopsubset_created %>%
select(PdKeyT, BLUE, GREEN, RED, SWIR1, SWIR2) %>%
pivot_longer(
cols = -PdKeyT
) %>%
mutate(color = case_when(name=="BLUE" ~ "blue",
name=="GREEN" ~ "green",
name=="RED" ~ "red",
name=="SWIR1" ~ "magenta",
name=="SWIR2" ~ "violet"))%>%
mutate(range = cut(PdKeyT, c(-Inf, -10, 30, Inf), c("Low", "Mid", "High"))) %>%
group_split(name)
p <- ggplot()
for (i in 1:5) p <- p geom_point(data=df_long_list[[i]], aes(value, PdKeyT, color=color, alpha=range))
geom_smooth(data=df_long_list[[i]], aes(value, PdKeyT, group=range), method = lm, se=TRUE)
theme(legend.position="none")
stat_cor(data=df_long_list[[i]], aes(value, PdKeyT,
label=paste("Spearman",..r.label.., ..p.label.., sep = "~`,`~")), method="spearman",
# label.x.npc="left", label.y.npc="top", hjust=0)
label.x = 3, label.y = 70)
stat_cor(data=df_long_list[[i]], aes(value, PdKeyT,
label=paste("Pearson",..r.label.., ..p.label.., sep = "~`,`~")), method="pearson",
# label.x.npc="left", label.y.npc="top", hjust=0)
label.x = 3, label.y = 65)
facet_grid(.~name, scales = "free")
theme_bw()
theme(panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
plot.margin = margin(120, 10, 120, 10),
panel.border = element_rect(fill = NA, color = "black"))
p
Here is how you could do it:
- select all relevant columns
- bring in long format
- add color column to dataframe
- make a list of dataframes with
group_split - use a for loop to iterate over each of the 5 dataframes in the list
- within the loop add
stat_corfor pearson and spearman fromggpubrpackage - facet and do some formatting
library(tidyverse)
library(ggpubr)
df_long_list <- loopsubset_created %>%
select(PdKeyT, BLUE, GREEN, RED, SWIR1, SWIR2) %>%
pivot_longer(
cols = -PdKeyT
) %>%
mutate(color = case_when(name=="BLUE" ~ "blue",
name=="GREEN" ~ "green",
name=="RED" ~ "red",
name=="SWIR1" ~ "magenta",
name=="SWIR2" ~ "violet"))%>%
group_split(name)
p <- ggplot()
for (i in 1:5) p <- p geom_point(data=df_long_list[[i]], aes(value, PdKeyT, color=color))
geom_smooth(data=df_long_list[[i]], aes(value, PdKeyT), method = lm, se=TRUE)
theme(legend.position="none")
stat_cor(data=df_long_list[[i]], aes(value, PdKeyT,
label=paste("Spearman",..r.label.., ..p.label.., sep = "~`,`~")), method="spearman",
# label.x.npc="left", label.y.npc="top", hjust=0)
label.x = 3, label.y = 70)
stat_cor(data=df_long_list[[i]], aes(value, PdKeyT,
label=paste("Pearson",..r.label.., ..p.label.., sep = "~`,`~")), method="pearson",
# label.x.npc="left", label.y.npc="top", hjust=0)
label.x = 3, label.y = 65)
facet_grid(.~name, scales = "free_y")
theme_bw()
theme(panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
plot.margin = margin(120, 10, 120, 10),
panel.border = element_rect(fill = NA, color = "black"))
p
CodePudding user response:
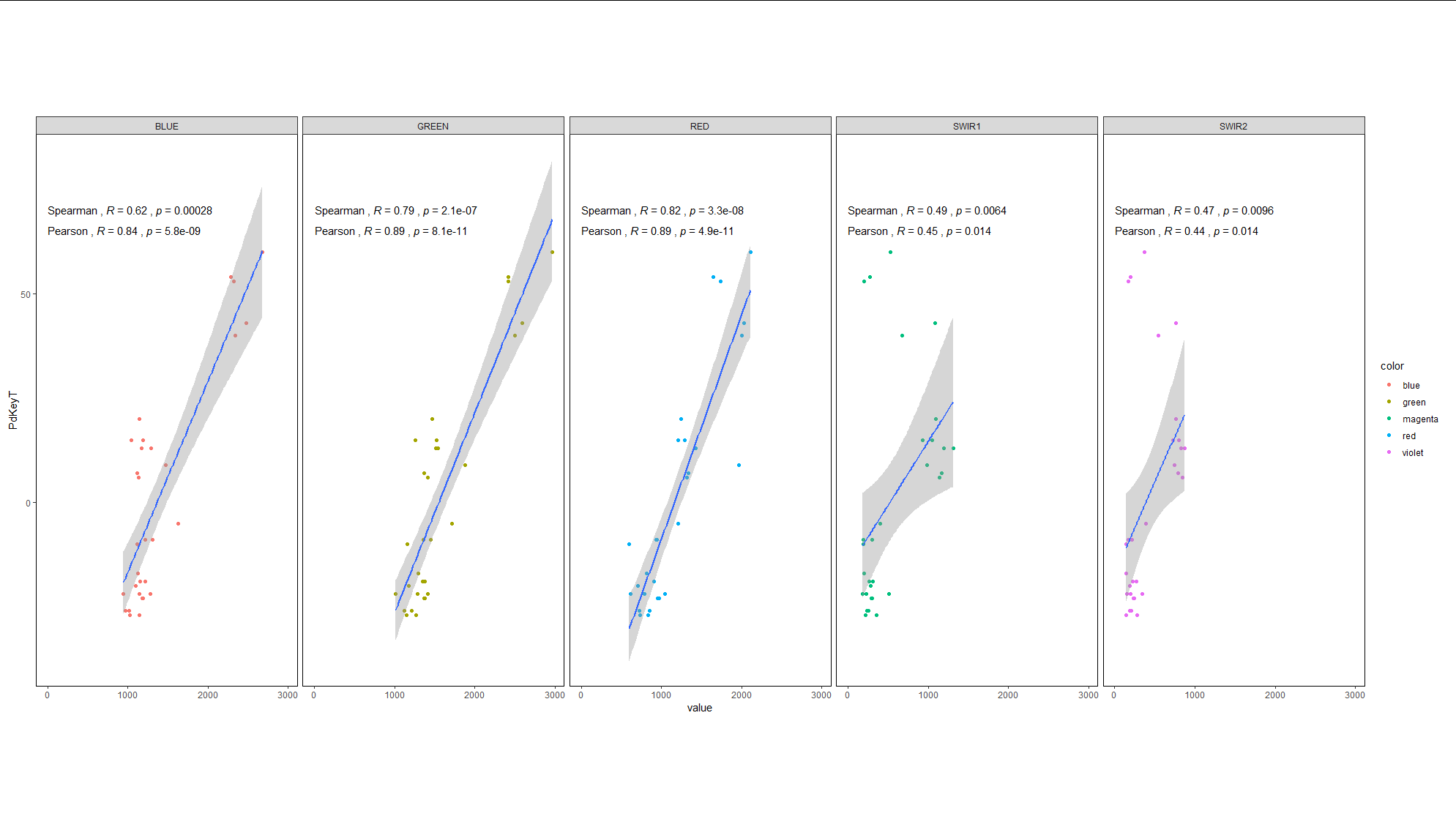
I think this fulfills most of your requirements, other than the correlation annotations. If, as you mention in your question, you wish to have 3 regressions per panel (one for each of the three ranges of PdkeyT) you would also need 3 correlation coefficients and p values per panel, which will be messy.
The reason why you have not seen tutorials for having different facets per variable is that this is not what facets are. Facets are a way of displaying data that have the same x and y axis but differ by some other categorical variable. They are not intended as a way of plotting different x variables against the same y variable. What you are describing is 5 distinct plots side-by-side, not facets.
Having said that, it is still possible to create the plot you are looking for with creative use of facets. You first need to shape the data into long format so that the values of the different x axis columns get stacked into a single column called value, and a new column called name is created to label each value according to which column it originally came from.
We can then use the new value column as our x axis variable, and facet according to the name column.
To make this look more authentic, we make some theme adjustments to ensure the facet strips resemble axis labels:
library(dplyr)
library(tidyr)
library(ggplot2)
loopsubset_created %>%
select(PdKeyT, BLUE, GREEN, RED, SWIR1, SWIR2) %>%
pivot_longer(-1) %>%
mutate(range = cut(PdKeyT, c(-Inf, -10, 30, Inf), c("Low", "Mid", "High"))) %>%
ggplot(aes(value, PdKeyT, color = name))
geom_point(aes(alpha = range))
geom_smooth(aes(group = range), size = 0.1,
method = "lm", formula = y ~ x, color = "black")
labs(x = "")
facet_grid(.~name, switch = "x", scales = "free_x")
scale_color_manual(values = c("blue", "green", "red", "magenta", "violet"))
theme_bw()
theme(strip.placement = "outside",
strip.background = element_blank(),
plot.margin = margin(120, 10, 120, 10),
legend.position = "none")
CodePudding user response:
To panel plots use facet_wrap or facet_grid. Also, generally ggplot2 works better when your data are in a long format. This allows you to assign a variable to an aesthetic rather than do it manually as you have.
library(ggplot2)
library(tidyr)
library(purrr)
library(dplyr)
library(tibble)
# lengthen your data so variable names are in a column
df <- loopsubset_created %>%
pivot_longer(cols = c(BLUE:RED, starts_with("SWIR")))
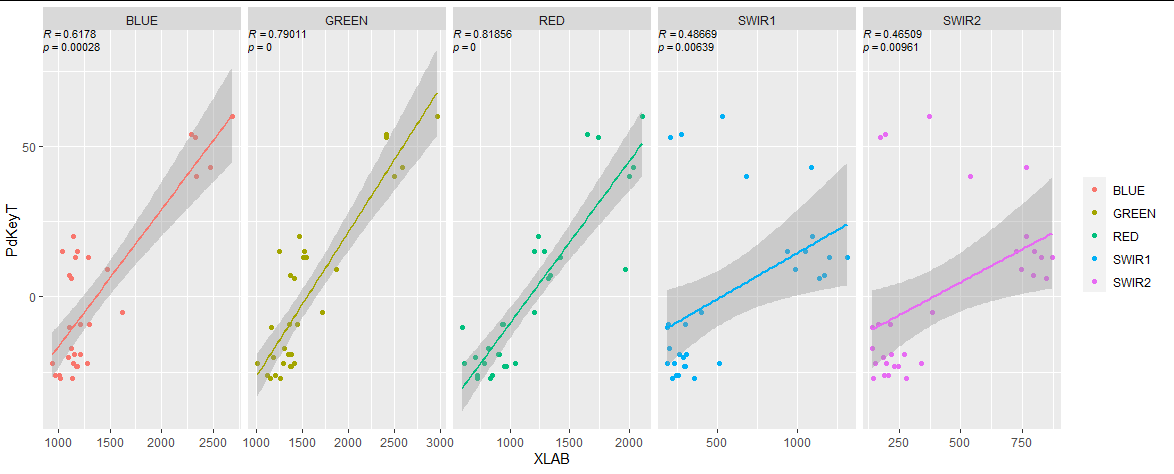
# get correlation coef and pvalue
r <- map(split(df, ~ name), ~ with(.x, c(cor(PdKeyT, value, method = "spearman"),
cor.test(PdKeyT, value, method = "spearman")$p.value))) %>%
bind_rows() %>%
rownames_to_column("i") %>% # first row is coef, second row is p value
pivot_longer(-i) %>%
mutate(lab = ifelse(i == 1,
# formatted so will be parsed by geom_text
sprintf("italic(R) == %0.5f", value),
sprintf("italic(p) == %0.5f", value)),
x = -Inf, # left of panel
y = Inf, # top of panel,
vjust = ifelse(i == 1, 0.75, 2)) # put p-value below
df %>%
ggplot(aes(x = value, y = PdKeyT, color = name))
geom_point()
geom_text(data = r,
aes(x = x, y = y,
label = lab,
vjust = vjust),
size = 3,
parse = T,
inherit.aes = F)
geom_smooth(method = "lm",
se = T,
formula = y ~ x,
show.legend = F)
facet_grid(~ name,
scales = "free_x")
labs(color = element_blank(),
x = "XLAB")