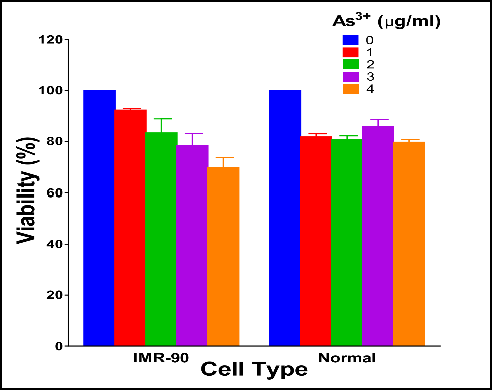
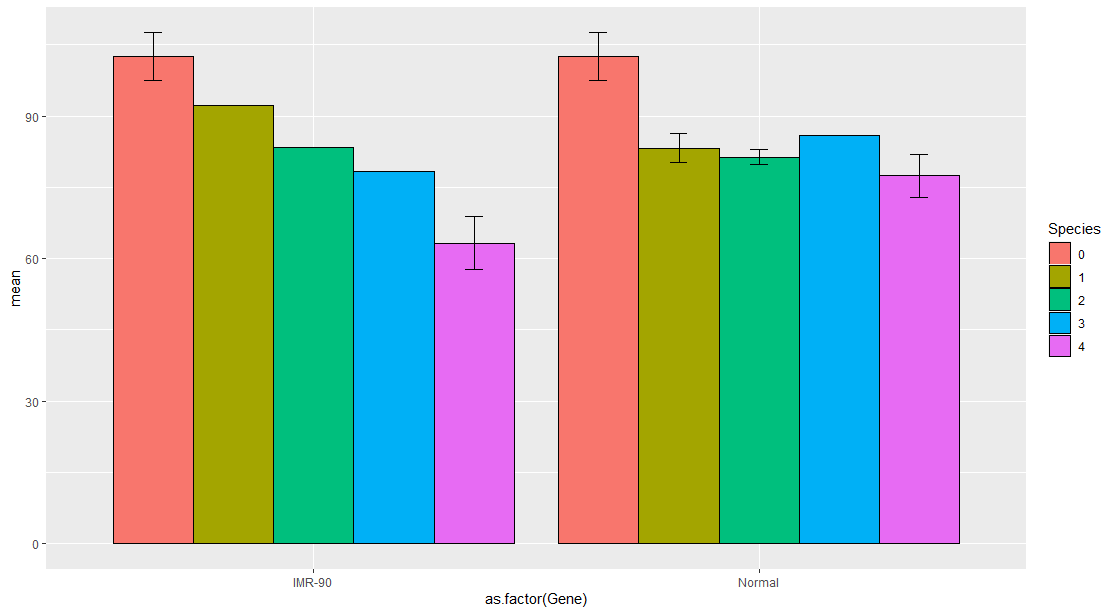
I am new to R and trying to build a grouped barchart that shows two groups for two different situations. I am a beginner so i'm trying to replicate from examples i've seen online.
#create dataframe
Gene<-c("IMR-90","Normal","IMR-90","Normal","IMR-90","Normal","IMR-90","Normal","IMR-90","Normal")
count1<-c(100,100,92.2453,81.76718,83.326,80.61812,78.34494,85.92033,69.75771,79.64783)
count2<-c(100,100,92.2453,81.76718,83.326,80.61812,78.34494,85.92033,69.75771,79.64783)
count3<-c(100,100,92.2453,81.76718,83.326,80.61812,78.34494,85.92033,69.75771,79.64783)
count4<-c(100,100,92.2453,81.76718,83.326,80.61812,78.34494,85.92033,69.75771,79.64783)
Species<-c("0","0","1","1","2","2","3","3","4","4")
cols = c(2,3,4,5,6,7,8,9,10,11)
df<-data.frame(Gene,count1,count2,count3,count4,Species)
df1 = transform(df, mean=rowMeans(df[cols]), sd=apply(df[cols],1, sd))
ggplot(df1, aes(x=as.factor(Gene), y=mean, fill=Species))
geom_bar(position=position_dodge(), stat="identity", colour='black')
geom_errorbar(aes(ymin=mean-sd, ymax=mean sd), width=.2,position=position_dodge(.9))
My R code is above and below is what I would like to achieve.
CodePudding user response:
It is advisable to check first where your error occurs as there is nothing wrong with your chart (ok perhaps not finished in all details but did not throw the error).
Your data preparation went wrong. It is also wise if you want to work with a standard deviation that you actually give us data that gives us a SD value not being 0 ;)
Gene<-c("IMR-90","Normal","IMR-90","Normal","IMR-90","Normal","IMR-90","Normal","IMR-90","Normal")
count1<-c(100,100,92.2453,81.76718,83.326,83.61812,78.34494,85.92033,62.75771,79.64783)
count2<-c(110,100,92.2453,87.76718,83.326,80.61812,78.34494,85.92033,65.75771,79.64783)
count3<-c(100,110,92.2453,81.76718,83.326,80.61812,78.34494,85.92033,55.75771,70.64783)
count4<-c(100,100,92.2453,81.76718,83.326,80.61812,78.34494,85.92033,68.75771,79.64783)
Species<-c("0","0","1","1","2","2","3","3","4","4")
library(data.table)
df<- data.table(Gene, count1, count2, count3, count4, Species)
cols <- c("count1", "count2", "count3", "count4")
df1 <- df[, .(mean = rowMeans(.SD), sd = sd(.SD)), by = .(Gene, Species), .SDcols = cols]
ggplot(df1, aes(x=as.factor(Gene), y=mean, fill=Species))
geom_bar(position=position_dodge(), stat="identity", colour='black')
geom_errorbar(aes(ymin=mean-sd, ymax=mean sd), width=.2,position=position_dodge(.9))
CodePudding user response:
It seems like your problem is found in the transformation that gives me error
To add mean and standard deviation to your data you can use the dplyr package and the function mutate. For calculating the mean and sd for every row you just need to add rowwise() to your pipeline
Gene<-c("IMR-90","Normal","IMR-90","Normal","IMR-90","Normal","IMR-90","Normal","IMR-90","Normal")
count1<-c(100,100,92.2453,81.76718,83.326,80.61812,78.34494,85.92033,69.75771,79.64783)
count2<-c(100,100,92.2453,81.76718,83.326,80.61812,78.34494,85.92033,69.75771,79.64783)
count3<-c(100,100,92.2453,81.76718,83.326,80.61812,78.34494,85.92033,69.75771,79.64783)
count4<-c(100,100,92.2453,81.76718,83.326,80.61812,78.34494,85.92033,69.75771,79.64783)
Species<-c("0","0","1","1","2","2","3","3","4","4")
cols = c(2,3,4,5,6,7,8,9,10,11)
df<-data.frame(Gene,count1,count2,count3,count4,Species)
df1 <- df %>% rowwise() %>% mutate(mean=(count1 count2 count3 count4)/3, stdev = sd(c(count1, count2, count3, count4)))
ggplot(df1, aes(x=as.factor(Gene), y=mean, fill=Species))
geom_bar(position=position_dodge(), stat="identity", colour='black')
geom_errorbar(aes(ymin=mean-stdev, ymax=mean stdev), width=.2,position=position_dodge(.9))
labs(title="title", x= "X axis name", y = "Y axis name")