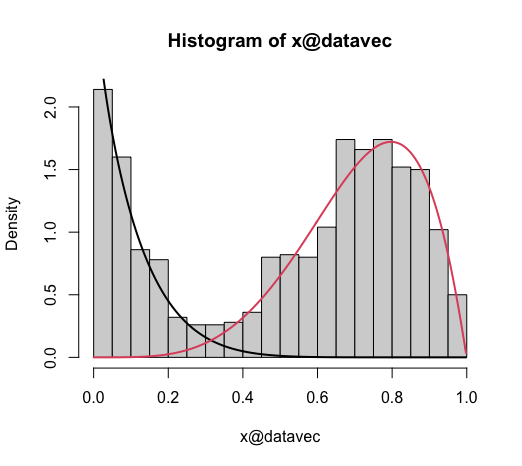
I am using a package called BetaMixture in R to fit a mixture of beta distributions for a data vector. The output is supplied to a hist that produces a good histogram with the mixture model components:
# Install and load the libraries
#install.packages("BetaModels")
library(BetaModels)
# Create a vector, fit mixture models and plot the histogram
vec <- c(rbeta(700, 5, 2), rbeta(300, 1, 10))
model <- BetaMixture(vec,2)
h <- hist(model, breaks = 35)
So far so good. Now how do I get this in ggplot? I inspected the h object but that is not different from the model object. They are exactly the same. I don't know how this hist even works for this class. What does it pull from the model to generate this plot other than the @datavec?
CodePudding user response:
You can get the hist function for BetaMixed objects using getMethod("hist", "BetaMixture").
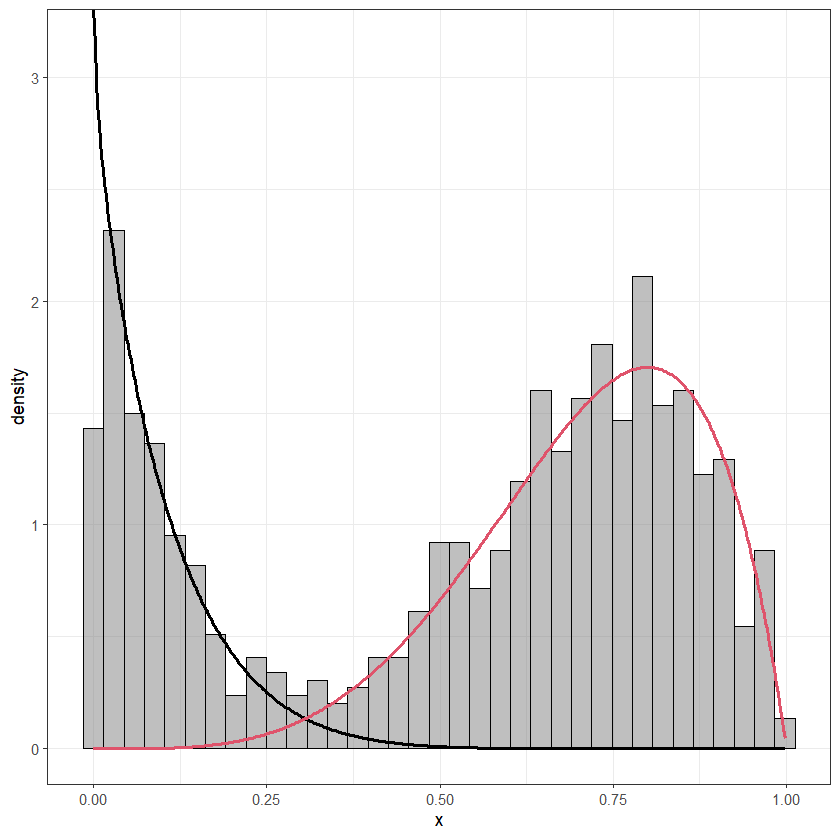
Below you can find a simple translation of this function into the "ggplot2 world".
myhist <- function (x, ...) {
.local <- function (x, mixcols = 1:7, breaks=25, ...)
{
df1 <- data.frame(x=x@datavec)
p <- ggplot(data=df1, aes(x=x))
geom_histogram(aes(y=..density..), bins=breaks, alpha=0.5, fill="gray50", color="black")
while (length(mixcols) < ncol(x@mle)) mixcols <- c(mixcols,
mixcols)
xv <- seq(0, 1, length = 502)[1:501]
for (J in 1:ncol(x@mle)) {
y <- x@phi[J] * dbeta(xv, x@mle[1, J], x@mle[2, J])
df2 <- data.frame(xv, y)
p <- p geom_line(data=df2, aes(xv, y), size=1, col=mixcols[J])
}
p <- p theme_bw()
invisible(p)
}
.local(x, ...)
}
library(ggplot2)
# Now p is a ggplot2 object.
p <- myhist(model, breaks=35)
print(p)
CodePudding user response:
The object returned by BetaMixture is a S4 class object with 2 slots that are of interest.
Slot Z returns a matrix of the probabilities of each data point belonging to each of the distributions.
So in the first 6 rows, all points belong to the 2nd distribution.
head(model@Z)
# [,1] [,2]
#[1,] 1.354527e-04 0.9998645
#[2,] 4.463074e-03 0.9955369
#[3,] 1.551999e-03 0.9984480
#[4,] 1.642579e-03 0.9983574
#[5,] 1.437047e-09 1.0000000
#[6,] 9.911427e-04 0.9990089
And slot mle return the maximum likelihood estimates of the parameters.
Now use those values to create a data.frame of the vector and a data.frame of the parameters.
df1 <- data.frame(vec)
df1$component <- factor(apply(model@Z, 1, which.max))
colors <- as.integer(levels(df1$component))
params <- as.data.frame(t(model@mle))
names(params) <- c("shape1", "shape2")
Plot the data.
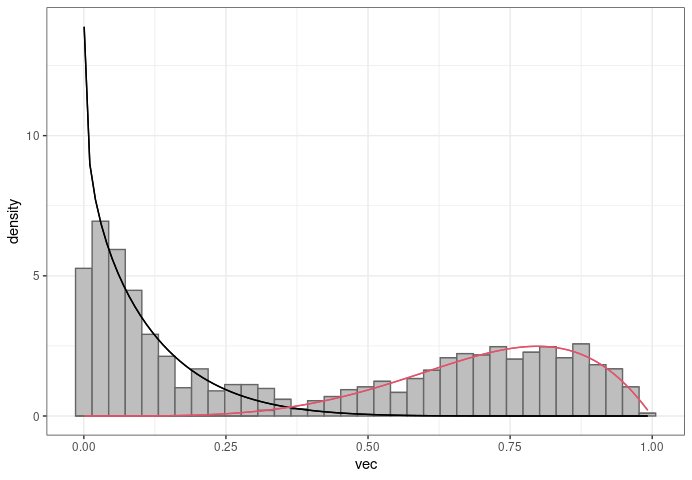
library(ggplot2)
g <- ggplot(df1, aes(x = vec, group = component))
geom_histogram(aes(y = ..density..),
bins = 35, fill = "grey", color = "grey40")
for(i in 1:nrow(params)){
sh1 <- params$shape1[i]
sh2 <- params$shape2[i]
g <- g stat_function(
fun = dbeta,
args = list(shape1 = sh1, shape2 = sh2),
color = colors[i]
)
}
suppressWarnings(print(g theme_bw()))