I m trying to find the mean of each row in a data frame for each group in columns. My input data is M, where the samples are grouped by annotation table:
M <- matrix(runif(40), nrow = 5, ncol = 8)
rownames(M) <- paste('Gene', seq(1:5), sep = "")
colnames(M) <-paste('Sample', seq(1:8), sep = "")
Annotation <- data.frame(Sample = paste('Sample', seq(1:8), sep = ""),Group = c(rep('A',2), rep('B', 2), rep('C',4)) )
Now I want to calculate the mean value for each rows in data frame for each group in annotation file and get another data frame/ or add to current data frame like:
The output values are random. Thank you.
CodePudding user response:
Update: Thanks to akrun! Here is a more automated version:
df %>%
pivot_longer(cols = -Gene, names_to = 'Sample') %>%
left_join(Annotation %>% mutate(Sample = str_remove(Sample, '[a-z] '), Group = str_c('Mean_Group_', Group))) %>%
dplyr::select(-Sample) %>%
pivot_wider(names_from = Group, values_from = value, values_fn = mean)
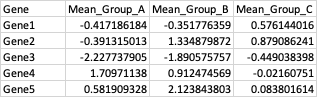
Gene Mean_Group_A Mean_Group_B Mean_Group_C
<chr> <dbl> <dbl> <dbl>
1 G1 -0.417 -0.352 0.576
2 G2 -0.391 1.33 0.879
3 G3 -2.23 -1.89 -0.449
4 G4 1.71 0.912 -0.0216
5 G5 0.582 2.12 0.0838
First answer:
One way would be to use pmap_dfr:
library(dplyr)
library(purrr)
df %>%
mutate(pmap_dfr(across(S1:S2),
~ data.frame(Mean_GroupA = mean(c(...))))) %>%
mutate(pmap_dfr(across(S3:S4),
~ data.frame(Mean_GroupB = mean(c(...))))) %>%
mutate(pmap_dfr(across(S5:S8),
~ data.frame(Mean_GroupC = mean(c(...)))))
Gene S1 S2 S3 S4 S5 S6 S7 S8 Mean_GroupA Mean_GroupB Mean_GroupC
1 G1 -2.9631013 2.128729 1.5440470 -2.2475997 -2.821792 2.789957 0.3067319 2.02967878 -0.4171862 -0.3517764 0.57614401
2 G2 -2.9881341 2.205504 0.1252083 2.5445515 2.419377 1.629860 -0.5366258 0.00373429 -0.3913150 1.3348799 0.87908623
3 G3 -2.3801595 -2.075316 -0.8376676 -2.9434839 2.162581 -1.089719 -0.5378263 -2.33118860 -2.2277379 -1.8905757 -0.44903839
4 G4 2.3143576 1.105065 1.2648060 0.5601431 -2.775243 1.145832 2.1268387 -0.58385730 1.7097114 0.9124746 -0.02160752
5 G5 -0.1625891 1.326408 1.4469643 2.8007233 2.057994 -0.041658 -2.0802055 0.39907598 0.5819093 2.1238438 0.08380163
data:
df <- structure(list(Gene = c("G1", "G2", "G3", "G4", "G5"), S1 = c(-2.9631013,
-2.9881341, -2.3801595, 2.31435765, -0.1625891), S2 = c(2.1287289,
2.20550407, -2.0753163, 1.10506511, 1.32640774), S3 = c(1.54404698,
0.12520826, -0.8376676, 1.26480602, 1.4469643), S4 = c(-2.2475997,
2.54455148, -2.9434839, 0.56014312, 2.8007233), S5 = c(-2.8217917,
2.41937685, 2.16258073, -2.7752431, 2.05799403), S6 = c(2.78995706,
1.62985958, -1.0897194, 1.14583159, -0.041658), S7 = c(0.30673189,
-0.5366258, -0.5378263, 2.12683874, -2.0802055), S8 = c(2.02967878,
0.00373429, -2.3311886, -0.5838573, 0.39907598)), class = "data.frame", row.names = c(NA,
-5L))
CodePudding user response:
If this is your data
da
X1 X2 X3 X4 X5
id S1 S2 S3 S4 S5
grp A A B B C
G1 0.6899627 0.1564079 1.1781793 0.8965239 0.8775281
G2 -1.1416318 0.3727000 1.3214322 -0.5519827 0.9628846
G3 -0.7996085 2.0349177 -1.7346331 1.7266299 1.3586755
G4 -1.00741747 -0.01393747 1.10712443 -0.23152309 -0.07544300
G5 -0.009610289 -1.109753618 0.687225281 0.237404982 0.388353962
X6 X7
id S6 S7
grp C D
G1 -2.1339055 -0.8832058
G2 0.6112205 0.4707905
G3 0.6396841 -0.5601753
G4 1.24123377 -0.36939366
G5 0.420034765 -0.288319580
you can extract the means like this
d <- data.frame( t(da) )
d
id grp G1 G2 G3 G4 G5
1 S1 A 0.6899627 -1.1416318 -0.7996085 -1.00741747 -0.009610289
2 S2 A 0.1564079 0.3727000 2.0349177 -0.01393747 -1.109753618
3 S3 B 1.1781793 1.3214322 -1.7346331 1.10712443 0.687225281
4 S4 B 0.8965239 -0.5519827 1.7266299 -0.23152309 0.237404982
5 S5 C 0.8775281 0.9628846 1.3586755 -0.07544300 0.388353962
6 S6 C -2.1339055 0.6112205 0.6396841 1.24123377 0.420034765
7 S7 D -0.8832058 0.4707905 -0.5601753 -0.36939366 -0.288319580
data.frame( t( aggregate( cbind( G1, G2, G3, G4, G5 ) ~ grp , d,
function(x) mean(as.numeric(x)) ) ) )
X1 X2 X3 X4
grp A B C D
G1 0.4231853 1.0373516 -0.6281887 -0.8832058
G2 -0.3844659 0.3847248 0.7870526 0.4707905
G3 0.6176546 -0.0040016 0.9991798 -0.5601753
G4 -0.5106775 0.4378007 0.5828954 -0.3693937
G5 -0.5596820 0.4623151 0.4041944 -0.2883196
Data
da <- structure(c("S1", "A", " 0.6899627", "-1.1416318", "-0.7996085",
"-1.00741747", "-0.009610289", "S2", "A", " 0.1564079", " 0.3727000",
" 2.0349177", "-0.01393747", "-1.109753618", "S3", "B", " 1.1781793",
" 1.3214322", "-1.7346331", " 1.10712443", " 0.687225281", "S4",
"B", " 0.8965239", "-0.5519827", " 1.7266299", "-0.23152309",
" 0.237404982", "S5", "C", " 0.8775281", " 0.9628846", " 1.3586755",
"-0.07544300", " 0.388353962", "S6", "C", "-2.1339055", " 0.6112205",
" 0.6396841", " 1.24123377", " 0.420034765", "S7", "D", "-0.8832058",
" 0.4707905", "-0.5601753", "-0.36939366", "-0.288319580"), .Dim = c(7L,
7L), .Dimnames = list(c("id", "grp", "G1", "G2", "G3", "G4",
"G5"), NULL))