I am trying to run a non-linear regression on a dataset whereby I would like to run a new regression for each group. The data frame is much like this one:
Date <- as.POSIXct(c("2021-05-25","2021-05-20", "2021-05-21","2021-05-22",
"2021-05-23","2021-05-24" ,"2021-05-25","2021-05-20", "2021-05-21","2021-05-22",
"2021-05-23","2021-05-24" ,"2021-05-25","2021-05-20", "2021-05-21","2021-05-22",
"2021-05-23","2021-05-24" ,"2021-05-25","2021-05-20", "2021-05-21","2021-05-22",
"2021-05-23","2021-05-24" ,"2021-05-25"))
Ts <- rnorm(25, mean=10, sd=0.5)
Exp_flux <- 3.5*exp((Ts-10)/10)
Collar <- as.factor(c("t1","t2","t3","t4","t5","t1","t2","t3","t4","t5","t1","t2","t3","t4",
"t5","t1","t2","t3","t4","t5","t1","t2","t3","t4","t5"))
df <- data.frame(Date,Collar,Ts,Exp_flux)
df
Date Collar Ts Exp_flux
1 2021-05-25 t1 9.596453 3.361570
2 2021-05-20 t2 8.870983 3.126334
3 2021-05-21 t3 10.011902 3.504168
4 2021-05-22 t4 10.480873 3.672418
5 2021-05-23 t5 10.264998 3.593989
6 2021-05-24 t1 10.196256 3.569368
7 2021-05-25 t2 9.523135 3.337014
8 2021-05-20 t3 10.315953 3.612349
9 2021-05-21 t4 9.510503 3.332801
10 2021-05-22 t5 10.300981 3.606945
11 2021-05-23 t1 10.788605 3.787187
12 2021-05-24 t2 10.226902 3.580323
13 2021-05-25 t3 9.005530 3.168683
14 2021-05-20 t4 10.752006 3.773351
15 2021-05-21 t5 9.335704 3.275051
16 2021-05-22 t1 9.345418 3.278234
17 2021-05-23 t2 10.034693 3.512164
18 2021-05-24 t3 10.754786 3.774401
19 2021-05-25 t4 9.655313 3.381415
20 2021-05-20 t5 10.670903 3.742872
21 2021-05-21 t1 8.986950 3.162801
22 2021-05-22 t2 10.441217 3.657883
23 2021-05-23 t3 10.446326 3.659753
24 2021-05-24 t4 10.550104 3.697931
25 2021-05-25 t5 10.442247 3.658260
My aim here is to run a separate regression of Exp_flux vs Ts for each collar type. I know I could separate the main dataset into subsets for each collar and perform each regression manually but in reality there are more than 20 collar types and I figured there must be a more efficient way to do this. I have tried using the nlsList function of the nlme package, which just gives an empty list or (in previous cases) the regression of only the first collar:
fit.collars <- nlsList(Exp_Flux ~ SRref*q^((Ts-10)/10)| Collar,
data=df, start=list(SRref=3, q=2), na.action = na.omit )
summary(fit.collars)
Error in class(val) <- c("summary.nlsList", class(val)) :
attempt to set an attribute on NULL
I must be using the nlsList function incorrectly but I can't figure out how so. Tutorials on this function are pretty scarce online. Can anyone advise on this or a relatively simple alternative?
CodePudding user response:
Let me quote help("nls"):
The default settings of nls generally fail on artificial “zero-residual” data problems.
If I add some white noise and fix the typo, I get successful fits.
set.seed(42)
Date <- as.POSIXct(c("2021-05-25","2021-05-20", "2021-05-21","2021-05-22",
"2021-05-23","2021-05-24" ,"2021-05-25","2021-05-20", "2021-05-21","2021-05-22",
"2021-05-23","2021-05-24" ,"2021-05-25","2021-05-20", "2021-05-21","2021-05-22",
"2021-05-23","2021-05-24" ,"2021-05-25","2021-05-20", "2021-05-21","2021-05-22",
"2021-05-23","2021-05-24" ,"2021-05-25"))
Ts <- rnorm(25, mean=10, sd=0.5)
Exp_flux <- 3.5*exp((Ts-10)/10) rnorm(25, sd = 0.01)
Collar <- as.factor(c("t1","t2","t3","t4","t5","t1","t2","t3","t4","t5","t1","t2","t3","t4",
"t5","t1","t2","t3","t4","t5","t1","t2","t3","t4","t5"))
df <- data.frame(Date,Collar,Ts,Exp_flux)
library(nlme)
fit.collars <- nlsList(Exp_flux ~ SRref*q^((Ts-10)/10)| Collar,
data=df, start=list(SRref=3, q=2), na.action = na.omit )
summary(fit.collars)
#works
Please consider carefully if you really want a pooled residual standard error.
CodePudding user response:
There are several problems:
- Exp_Flux in the formula differs from Exp_flux as the column name
- The question uses random numbers without set.seed so the data is not reproducible. We have used the data shown in the Note at the end for reproducibility.
- Better starting values may be required. Using the data in the Note at the end the starting values in the question work as is but since the data is not reproducible we have added better starting valu3es just in case they do not in the actual data.
- Add control = nls.control(scaleOffset = 1) argument to handle zero residuals. Note that scaleOffset was introduced in R 4.1.2 and is not available in earlier versions of R.
Code--
library(nlme)
# get starting values
fit0 <- lm(log(Exp_flux) ~ I((Ts-10)/10), df)
st <- setNames(exp(coef(fit0)), c("SRref", "q"))
fo2 <- Exp_flux ~ SRref * q^((Ts-10)/10) | Collar
fit2 <- nlsList(fo2, data=df, start = st, na.action = na.omit,
control = nls.control(scaleOffset = .1))
fit2
giving:
Call:
Model: Exp_flux ~ SRref * q^((Ts - 10)/10) | Collar
Data: df
Coefficients:
SRref q
t1 3.5 2.718282
t2 3.5 2.718282
t3 3.5 2.718282
t4 3.5 2.718282
t5 3.5 2.718282
Degrees of freedom: 25 total; 15 residual
Residual standard error: 1.152352e-15
Grouped vs ungrouped
Note that for this data grouping by Collar is not significant. We can already observe that from the coefficients being the same but if that is not the case with the real data this is how to perform the test using anova.
# ungrouped
fo3 <- Exp_flux ~ SRref * q^((Ts-10)/10)
fit3 <- nls(fo3, data = df, start = st,
na.action = na.omit, control = list(scaleOffset = 1))
# grouped
fo4 <- Exp_flux ~ SRref[Collar] * q[Collar]^((Ts-10)/10)
fit4 <- nls(fo4, data = df,
start = list(SRref = rep(st[[1]], 5), q = rep(st[[2]], 5)),
na.action = na.omit, control = list(scaleOffset = 1))
anova(fit3, fit4)
giving:
Analysis of Variance Table
Model 1: Exp_flux ~ SRref * q^((Ts - 10)/10)
Model 2: Exp_flux ~ SRref[Collar] * q[Collar]^((Ts - 10)/10)
Res.Df Res.Sum Sq Df Sum Sq F value Pr(>F)
1 23 1.9919e-29
2 15 1.9919e-29 8 0 0 1
Linear model
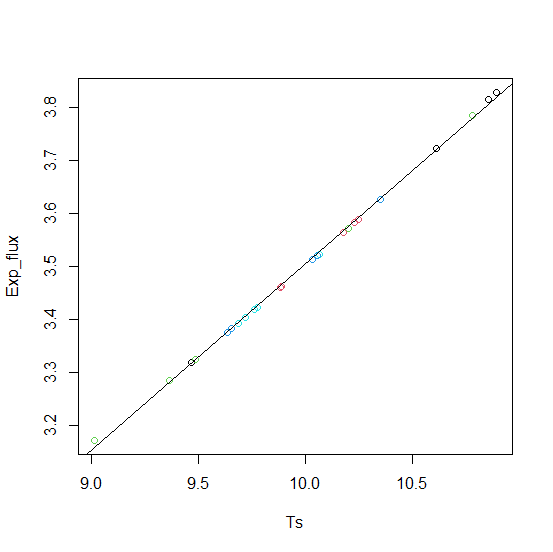
Note that a simple linear model fits the data quite well.
plot(Exp_flux ~ Ts, df, col = df$Collar)
fm0 <- lm(Exp_flux ~ Ts, df)
abline(fm0)
Note
We used this data:
set.seed(123)
Date <- as.POSIXct(c("2021-05-25","2021-05-20", "2021-05-21","2021-05-22",
"2021-05-23","2021-05-24" ,"2021-05-25","2021-05-20", "2021-05-21","2021-05-22",
"2021-05-23","2021-05-24" ,"2021-05-25","2021-05-20", "2021-05-21","2021-05-22",
"2021-05-23","2021-05-24" ,"2021-05-25","2021-05-20", "2021-05-21","2021-05-22",
"2021-05-23","2021-05-24" ,"2021-05-25"))
Ts <- rnorm(25, mean=10, sd=0.5)
Exp_flux <- 3.5*exp((Ts-10)/10)
Collar <- as.factor(c("t1","t2","t3","t4","t5","t1","t2","t3","t4","t5","t1","t2","t3","t4",
"t5","t1","t2","t3","t4","t5","t1","t2","t3","t4","t5"))
df <- data.frame(Date,Collar,Ts,Exp_flux)
Update 3
There was an error in copying the formula from the question in the prior version of this answer as well as a change made to make it work. Have fixed those now and it now works using (1) an improvement in starting value whichi may or may not be needed and (2) the addition of the scaleOffset parameter. @Roland pointed out that the model was wrong and that the model has zero residuals.
Also added sections on comparing grouped vs ungrouped and on using a simple linear model.