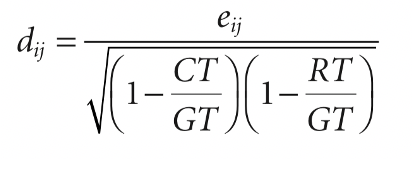I have a cross-tab (dataframe format), from which I have calculated the chi-sq standardized residuals. Below I provide the two reproducible datasets.
Cross-tab:
df <- structure(c(310, 36, 0, 0, 212, 158, 9, 0, 21, 35, 17, 4, 25,
102, 49, 18, 7, 35, 51, 28), .Dim = 4:5, .Dimnames = list(c("none",
"grade1", "grade2", "grade3"), c("0-9", "10-19", "20-29", "30-39",
"40 ")))
Standardized residuals
st.residuals <- structure(c(9.882, -7.267, -6.247, -3.935, 1.21, 3.035, -5.162,
-4.119, -2.96, 1.945, 2.821, 0.298, -7.492, 4.82, 5.796, 3.161,
-7.005, -0.738, 10.11, 9.704), .Dim = 4:5, .Dimnames = list(c("none",
"grade1", "grade2", "grade3"), c("0-9", "10-19", "20-29", "30-39",
"40 ")))
Goal
what I am after is to calculate the adjusted standardized residuals, which entails dividing each standardized residual by the quantity indicated in the below pict, where GT is the table grand total, CT is the column total, and RC is the row total:
Where I am stuck
I am having hard time in figuring out (my bad) how to implement in R the calculation for the denominator. In particular, I do not know how to code so that for each cell R will take into account the corresponding row and column total.
CodePudding user response:
1) R already has this in chisq.test:
chisq.test(df)$stdres
2) or the following. residuals is the same as st.residuals in the question and the final line produces the same result as the line above.
expected <- outer(rowSums(df), colSums(df)) / sum(df)
residuals <- (df - expected) / sqrt(expected)
residuals / sqrt(outer((1 - rowSums(df) / sum(df)), (1 - colSums(df) / sum(df))))
3) Alternately we can use sweep to calculate (1) above. residuals is from (2) and, as mentioned, equals st.residuals in the question.
residuals |>
sweep(1, sqrt(1 - rowSums(df) / sum(df)), `/`) |>
sweep(2, sqrt(1 - colSums(df) / sum(df)), `/`)