I have three arrays that essentially correspond to a matrix of gene expression values and then column labels specifying condition IDs and row values specifying a specific gene. I'm trying to define a function that will plot a histogram by just providing the gene name.
Basically I need to specify YAL001C and create a histogram of the values across the row. I'm very new to matplotlib and I'm not sure how do this. Would it have something to do with using something like an np.where(gene = YAL001C) argument? I guess I'm just not sure where that would fit into code for matplotlib.
I currently have the following code, but it doesn't work:
def histogram(gene):
fig = plt.figure()
ax = fig.add_subplot(1, 1, 1)
x = np.where(geneList == gene, exprMat)
bins = 50
ax.hist(x, bins, color = 'green', edgecolor = 'black', alpha = 0.8 )
plt.show()
CodePudding user response:
Here is one of the ways you could accomplish that (passing the data related to the corresponding row to the method):
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
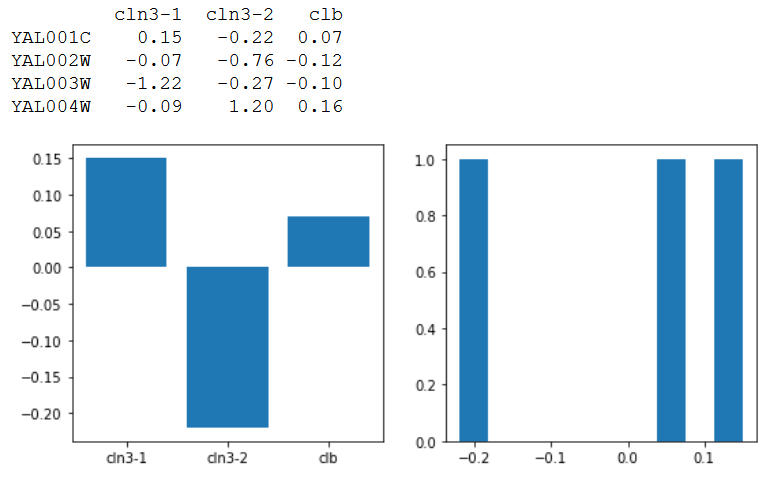
data = np.array([[0.15, -0.22, 0.07],
[-0.07, -0.76, -0.12],
[-1.22, -0.27, -0.1],
[-0.09, 1.2, 0.16]
])
df = pd.DataFrame(data=data,
index=['YAL001C', 'YAL002W', 'YAL003W', 'YAL004W'],
columns=['cln3-1', 'cln3-2', 'clb'])
print(df)
def plot_hist(gene):
fig, ax = plt.subplots(1,2, figsize=(9,4))
ax[0].bar(df.columns, df.loc[gene])
ax[1].hist(df.loc[gene])
plt.show()
plot_hist('YAL001C')
Left: bar-plot, Right: histogram
CodePudding user response:
In case you want to avoid using pandas, you can still accomplish what you want using numpy, but you need to add some codes to figure out what row corresponds to a given gene. Here is one of the ways you could code it:
import numpy as np
import matplotlib.pyplot as plt
data = np.array([[0.15, -0.22, 0.07],
[-0.07, -0.76, -0.12],
[-1.22, -0.27, -0.1],
[-0.09, 1.2, 0.16]
])
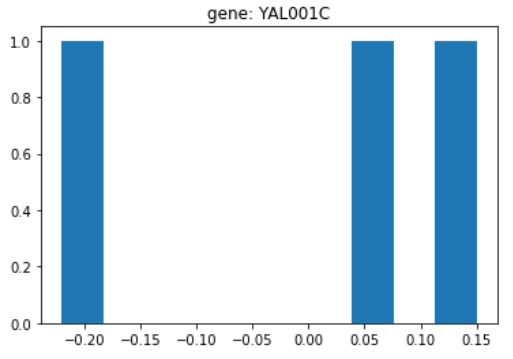
def plot_hist(gene):
list_genes = ['YAL001C', 'YAL002W', 'YAL003W', 'YAL004W']
if gene in list_genes:
sn_gene = list_genes.index(gene)
else:
print(f'{gene} is not in the list of genes')
return
fig, ax = plt.subplots(figsize=(6,4))
plt.hist(data[sn_gene,:])
plt.title(f'gene: {gene}')
plt.show()
plot_hist('YAL001C')