when we fit a linear model in like this,
x <- c(1.52, 1.6, 1.68, 1.75, 1.83)
y <- c(1.69, 1.74, 1.80, 1.93, 2.0)
fit <- lm(y ~ x)
summary(fit)
Output:
Call:
lm(formula = y ~ x)
Residuals:
1 2 3 4 5
0.021416 -0.012387 -0.036190 0.020482 0.006679
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.07633 0.19447 0.392 0.72093
x 1.04754 0.11579 9.047 0.00285 **
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 0.0282 on 3 degrees of freedom
Multiple R-squared: 0.9646, Adjusted R-squared: 0.9529
F-statistic: 81.85 on 1 and 3 DF, p-value: 0.002852
So, what I want is a latex output of this "exact" table for Rmarkdown pdf output file. I do not want such output that is produced that changes the table or does not include some information from this table. As I mean, exactly this same output but in latex output.
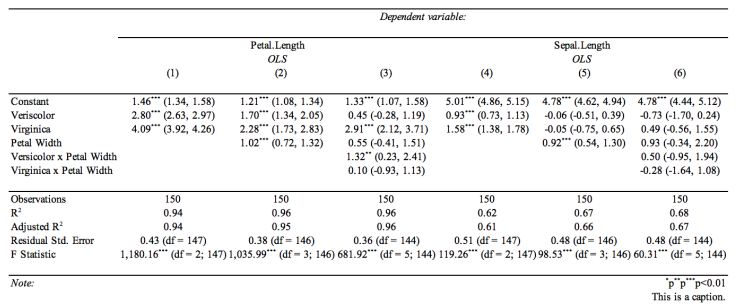
An example output would be like this image but for single regrassion model and also contains all the information generated by summary(lm(y~x))
Note: I have used such libraties sjPlot, stargazer, texreg etc.
Currently I am using R version 4.1.2
CodePudding user response:
Using echo=FALSE produces what you want.
---
output: pdf_document
---
```{r, echo=FALSE}
x <- c(1.52, 1.6, 1.68, 1.75, 1.83)
y <- c(1.69, 1.74, 1.80, 1.93, 2.0)
fit <- lm(y ~ x)
summary(fit)
```
CodePudding user response:
I am a big fan of modelsummary for making tables
library(tidyverse)
library(modelsummary)
library(broom)
library(kableExtra)
df = tibble(x = c(1.52, 1.6, 1.68, 1.75, 1.83),
y = 1.69, 1.74, 1.80, 1.93, 2.0 )
fit <- lm(y ~ x , data = df)
tidy(fit)
modelsummary(fit,
output = "kableExtra")
Which will produce a very basic LaTeX table. However, if you want to group them by dvs you can do something like this where you estimate your models, then use broom, and then
data("iris")
iris = iris %>%
mutate(versicolor = ifelse(Species == "versicolor", 1, 0),
virginica = ifelse(Species == "virginica", 1, 0))
## pass it to tidy for each model
tidy(model1)
modelsummary(list( model1 , model2, model3,
model4, model5, model6),
output = "kableExtra") %>%
add_header_above(c(" " = 1 , "Petal Length" = 3 , "Sepal Width" = 3))
Which should give you close to what you are looking for! You may need to look at more options in kableExtra.