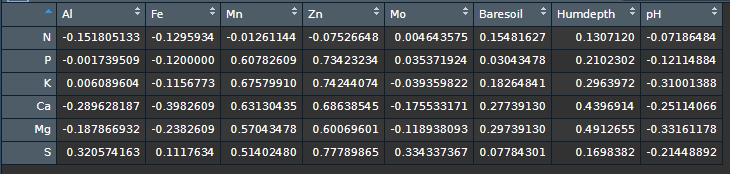
I need to calculate the correlation of some specific variables (columns).
To calculate the correlation of specific columns I get through this code:
df<-read.csv("http://renatabrandt.github.io/EBC2015/data/varechem.csv", row.names=1)
cor_df<-(cor(df, method="spearman")[1:6, 7:14])%>%as.data.frame()
output
However I would like R to create a new matrix but only with the correlations with a level of significance, whose p-value <0.05, only for the set [1:6, 7:14], that is to say exclude those not significant (p-value >0.05)
I expect the non-significant ones to be deleted, or filled in with NA, or a new data.frame with just the signifiers.
my expectavie is:
CodePudding user response:
Please find below one possible solution using Hmisc, corrplot and dplyr libraries
Reprex
- Computes the correlation coefficients and corresponding pvalues using the
rcorr()function of theHmisclibrary
library(Hmisc)
library(corrplot)
library(dplyr)
coeffs <- rcorr(as.matrix(df), type="spearman")[[1]][1:6, 7:14]
coeffs
#> Al Fe Mn Zn Mo Baresoil
#> N -0.151805133 -0.1295934 -0.01261144 -0.07526648 0.004643575 0.15481627
#> P -0.001739509 -0.1200000 0.60782609 0.73423234 0.035371924 0.03043478
#> K 0.006089604 -0.1156773 0.67579910 0.74244074 -0.039359822 0.18264841
#> Ca -0.289628187 -0.3982609 0.63130435 0.68638545 -0.175533171 0.27739130
#> Mg -0.187866932 -0.2382609 0.57043478 0.60069601 -0.118938093 0.29739130
#> S 0.320574163 0.1117634 0.51402480 0.77789865 0.334337367 0.07784301
#> Humdepth pH
#> N 0.1307120 -0.07186484
#> P 0.2102302 -0.12114884
#> K 0.2963972 -0.31001388
#> Ca 0.4396914 -0.25114066
#> Mg 0.4912655 -0.33161178
#> S 0.1698382 -0.21448892
pvalues <- rcorr(as.matrix(df), type="spearman")[[3]][1:6, 7:14]
pvalues
#> Al Fe Mn Zn Mo Baresoil
#> N 0.4788771 0.54615126 0.9533606683 7.266830e-01 0.9828194 0.4700940
#> P 0.9935636 0.57648987 0.0016290786 4.418653e-05 0.8696630 0.8877339
#> K 0.9774704 0.59039698 0.0002896520 3.264276e-05 0.8551122 0.3929703
#> Ca 0.1698232 0.05391473 0.0009388912 2.126270e-04 0.4119734 0.1894124
#> Mg 0.3793530 0.26221751 0.0036070461 1.909894e-03 0.5798929 0.1581543
#> S 0.1266908 0.60311127 0.0101838168 7.669395e-06 0.1103062 0.7176938
#> Humdepth pH
#> N 0.54266218 0.7386046
#> P 0.32412825 0.5728181
#> K 0.15961613 0.1404062
#> Ca 0.03156073 0.2365150
#> Mg 0.01477451 0.1134202
#> S 0.42754109 0.3141949
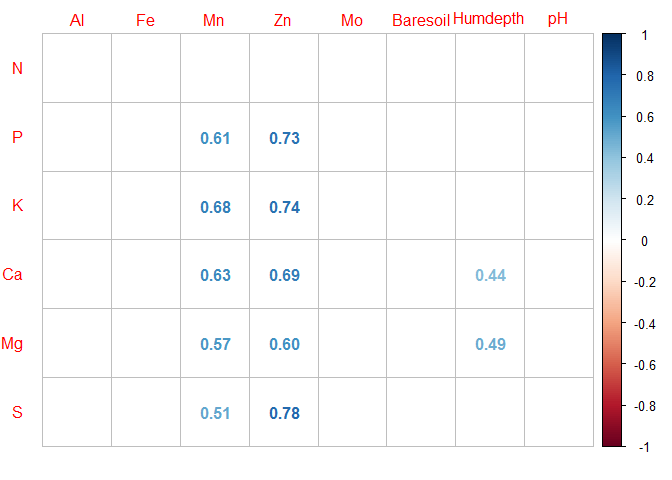
- Visualization using the
corrplot()function
r <- corrplot(coeffs,
method = "number",
p.mat = pvalues,
sig.level = 0.05, # displays only corr. coeff. for p < 0.05
insig = "blank", # else leave the cell blank
tl.srt = 0, # control the orintation of text labels
tl.offset = 1) # control of the offset of the text labels
- Use the results of the
corrplot()function to build a more "traditionnally" matrix of results
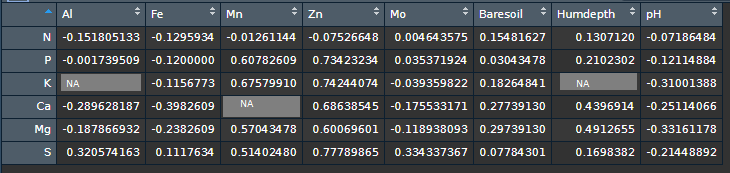
# Keep only the correlation coefficients for pvalues < 0.05
ResultsMatrix <- r$corrPos %>%
mutate(corr = ifelse(p.value < 0.05, corr, NA))
# Set factors to control the order of rows and columns in the final cross-table
ResultsMatrix$xName <- factor(ResultsMatrix$xName,
levels = c("Al", "Fe", "Mn", "Zn", "Mo", "Baresoil", "Humdepth", "pH"))
ResultsMatrix$yName <- factor(ResultsMatrix$yName,
levels = c("N", "P", "K", "Ca", "Mg", "S"))
# Build the cross-table and get a dataframe as final result
xtabs(corr ~ yName xName,
data = ResultsMatrix,
sparse = TRUE,
addNA = TRUE) %>%
as.matrix() %>%
as.data.frame()
- Output
#> Al Fe Mn Zn Mo Baresoil Humdepth pH
#> N NA NA NA NA NA NA NA NA
#> P NA NA 0.6078261 0.7342323 NA NA NA NA
#> K NA NA 0.6757991 0.7424407 NA NA NA NA
#> Ca NA NA 0.6313043 0.6863854 NA NA 0.4396914 NA
#> Mg NA NA 0.5704348 0.6006960 NA NA 0.4912655 NA
#> S NA NA 0.5140248 0.7778986 NA NA NA NA
Created on 2021-12-21 by the reprex package (v2.0.1)