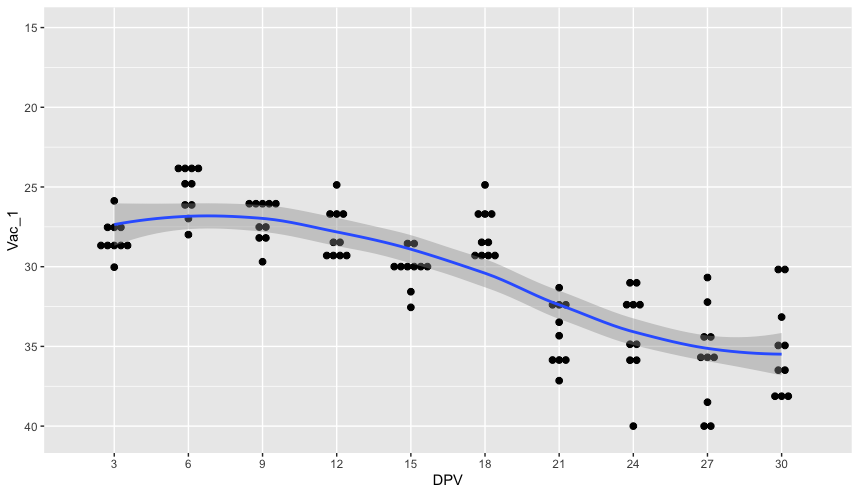
I have been reading a lot of the post associated to geom_smooth() and I think this is a new question, if not please link the solution and I will read that one!
I have a data set we are comparing qPCR detection results over the course of 30 days
ggplot(Vaccine1, aes(x= factor(DPV),y = VAC1))
geom_dotplot(binaxis="y", stackdir="center",dotsize = .5)
scale_y_reverse()
expand_limits(y=c(15,40))
This gives me the graph want, the detection trend/sampling day
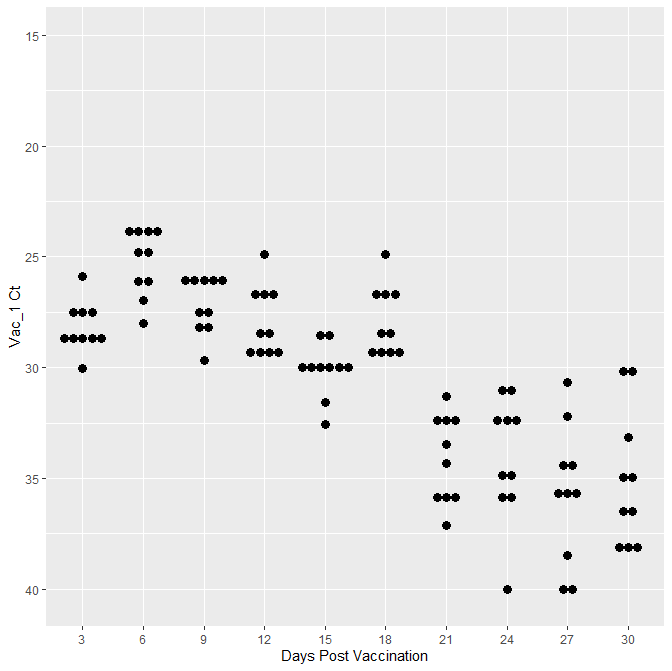
When I call the above code with geom_smooth() I get the same plot with out the line.
ggplot(Vaccine1, aes(x= factor(DPV),y = VAC1))
geom_dotplot(binaxis="y", stackdir="center",dotsize = .5)
scale_y_reverse()
expand_limits(y=c(15,40))
geom_smooth()
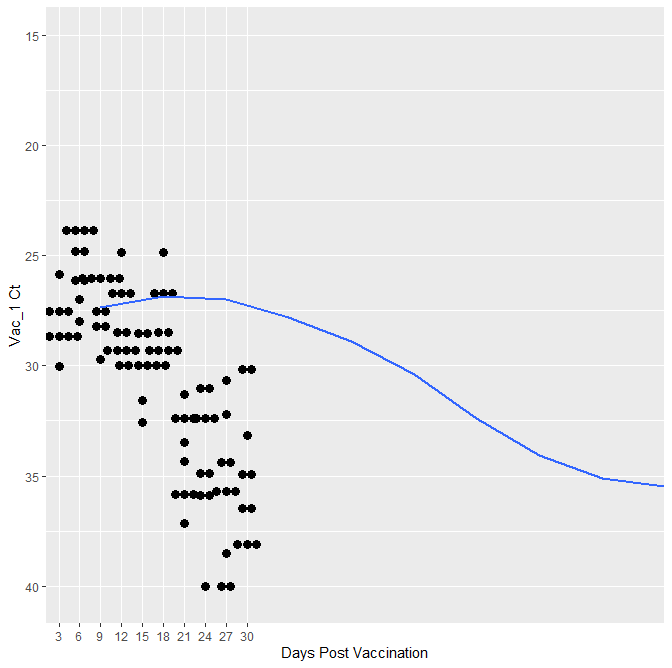
So after reading several different post I added this to the code
geom_smooth(method = "loess",aes(x=DPV,y=Vac_1),inherit.aes=FALSE,se=FALSE)
which then creates a plot with the data smashed to the left
It is the correct trend line, however no the visual I was going for. I think the issue has something to due with the initial ggplot(Vaccine1, aes(x= factor(DPV),y = VAC1)) becasue when I remove the factor I get the trend line over the data, but I do not get the per day results.
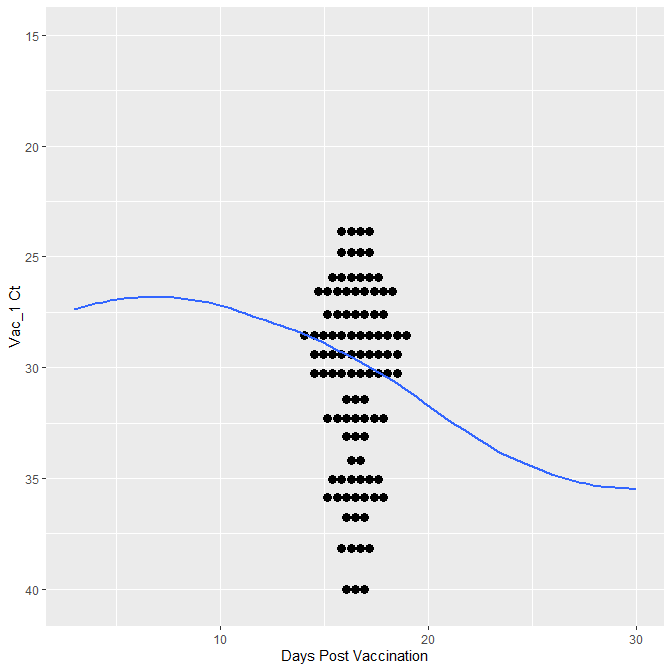
Does anyone have any thoughts on how I might correct this to get the trendline to overlay the individual day data? I am sure its a syntax issue that I am not seeing and if your fresh eyes can spot it please let me know!
Thank you in advance for your help.
EDIT Please find the blinded data here for your reference
Tag #,Vac_1,DPV
1,28.65,3
2,28.83,3
3,27.67,3
4,28.51,3
5,25.87,3
6,28.75,3
7,27.24,3
8,27.82,3
9,30.03,3
10,28.56,3
11,24.24,6
12,23.66,6
13,26.98,6
14,26.14,6
15,24.52,6
16,26.11,6
17,27.99,6
18,23.78,6
19,25.08,6
20,23.43,6
21,29.69,9
22,28.5,9
23,27.43,9
24,27.59,9
25,25.85,9
26,26.11,9
27,26.18,9
28,25.7,9
29,27.89,9
30,26.4,9
31,26.78,12
32,24.87,12
33,28.39,12
34,29.51,12
35,28.55,12
36,26.61,12
37,29.09,12
38,26.64,12
39,29.37,12
40,29.38,12
41,29.74,15
42,32.55,15
43,30.25,15
44,29.99,15
45,29.88,15
46,30.14,15
47,31.57,15
48,28.25,15
49,28.85,15
50,30.17,15
51,26.78,18
52,24.87,18
53,28.39,18
54,29.51,18
55,28.55,18
56,26.61,18
57,29.09,18
58,26.64,18
59,29.37,18
60,29.38,18
61,37.15,21
62,31.32,21
63,34.33,21
64,33.48,21
65,32.2,21
66,32.58,21
67,32.45,21
68,36.02,21
69,35.69,21
70,35.86,21
71,32.05,24
72,30.6,24
73,32.72,24
74,32.49,24
75,34.71,24
76,40,24
77,35.46,24
78,35.02,24
79,31.42,24
80,36.27,24
81,38.5,27
82,35.42,27
83,34.75,27
84,40,27
85,32.22,27
86,34.06,27
87,35.95,27
88,35.92,27
89,30.68,27
90,40,27
91,38.21,30
92,30.29,30
93,33.16,30
94,37.81,30
95,38.44,30
96,34.69,30
97,35.19,30
98,36.39,30
99,30.06,30
100,36.59,30
CodePudding user response:
x_brk = unique(Vaccine1$DPV)
ggplot(Vaccine1, aes(x= DPV, y = Vac_1))
geom_dotplot(binaxis="y", stackdir="center",dotsize = .5, aes(group = DPV))
scale_y_reverse()
scale_x_continuous(breaks = x_brk, minor_breaks = NULL)
expand_limits(y=c(15,40))
geom_smooth()
data
Vaccine1 <- data.frame(
Tag = c(1L,2L,3L,4L,5L,6L,7L,8L,9L,10L,
11L,12L,13L,14L,15L,16L,17L,18L,19L,20L,21L,22L,23L,
24L,25L,26L,27L,28L,29L,30L,31L,32L,33L,34L,35L,36L,
37L,38L,39L,40L,41L,42L,43L,44L,45L,46L,47L,48L,
49L,50L,51L,52L,53L,54L,55L,56L,57L,58L,59L,60L,61L,
62L,63L,64L,65L,66L,67L,68L,69L,70L,71L,72L,73L,
74L,75L,76L,77L,78L,79L,80L,81L,82L,83L,84L,85L,86L,
87L,88L,89L,90L,91L,92L,93L,94L,95L,96L,97L,98L,99L,
100L),
Vac_1 = c(28.65,28.83,27.67,28.51,25.87,28.75,
27.24,27.82,30.03,28.56,24.24,23.66,26.98,26.14,24.52,
26.11,27.99,23.78,25.08,23.43,29.69,28.5,27.43,27.59,
25.85,26.11,26.18,25.7,27.89,26.4,26.78,24.87,28.39,
29.51,28.55,26.61,29.09,26.64,29.37,29.38,29.74,32.55,
30.25,29.99,29.88,30.14,31.57,28.25,28.85,30.17,26.78,
24.87,28.39,29.51,28.55,26.61,29.09,26.64,29.37,29.38,
37.15,31.32,34.33,33.48,32.2,32.58,32.45,36.02,35.69,35.86,
32.05,30.6,32.72,32.49,34.71,40,35.46,35.02,31.42,
36.27,38.5,35.42,34.75,40,32.22,34.06,35.95,35.92,30.68,
40,38.21,30.29,33.16,37.81,38.44,34.69,35.19,36.39,
30.06,36.59),
DPV = c(3L,3L,3L,3L,3L,3L,3L,3L,3L,3L,6L,
6L,6L,6L,6L,6L,6L,6L,6L,6L,9L,9L,9L,9L,9L,9L,9L,
9L,9L,9L,12L,12L,12L,12L,12L,12L,12L,12L,12L,12L,
15L,15L,15L,15L,15L,15L,15L,15L,15L,15L,18L,18L,
18L,18L,18L,18L,18L,18L,18L,18L,21L,21L,21L,21L,21L,
21L,21L,21L,21L,21L,24L,24L,24L,24L,24L,24L,24L,24L,
24L,24L,27L,27L,27L,27L,27L,27L,27L,27L,27L,27L,
30L,30L,30L,30L,30L,30L,30L,30L,30L,30L)
)