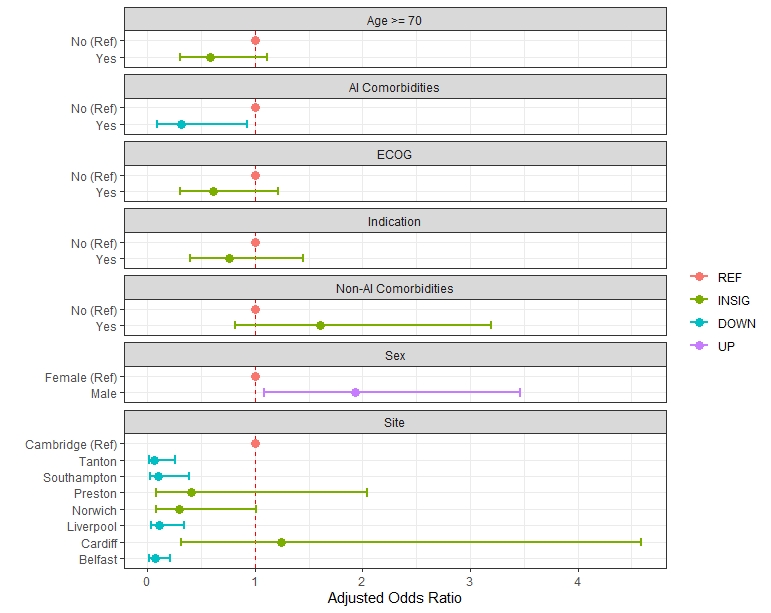
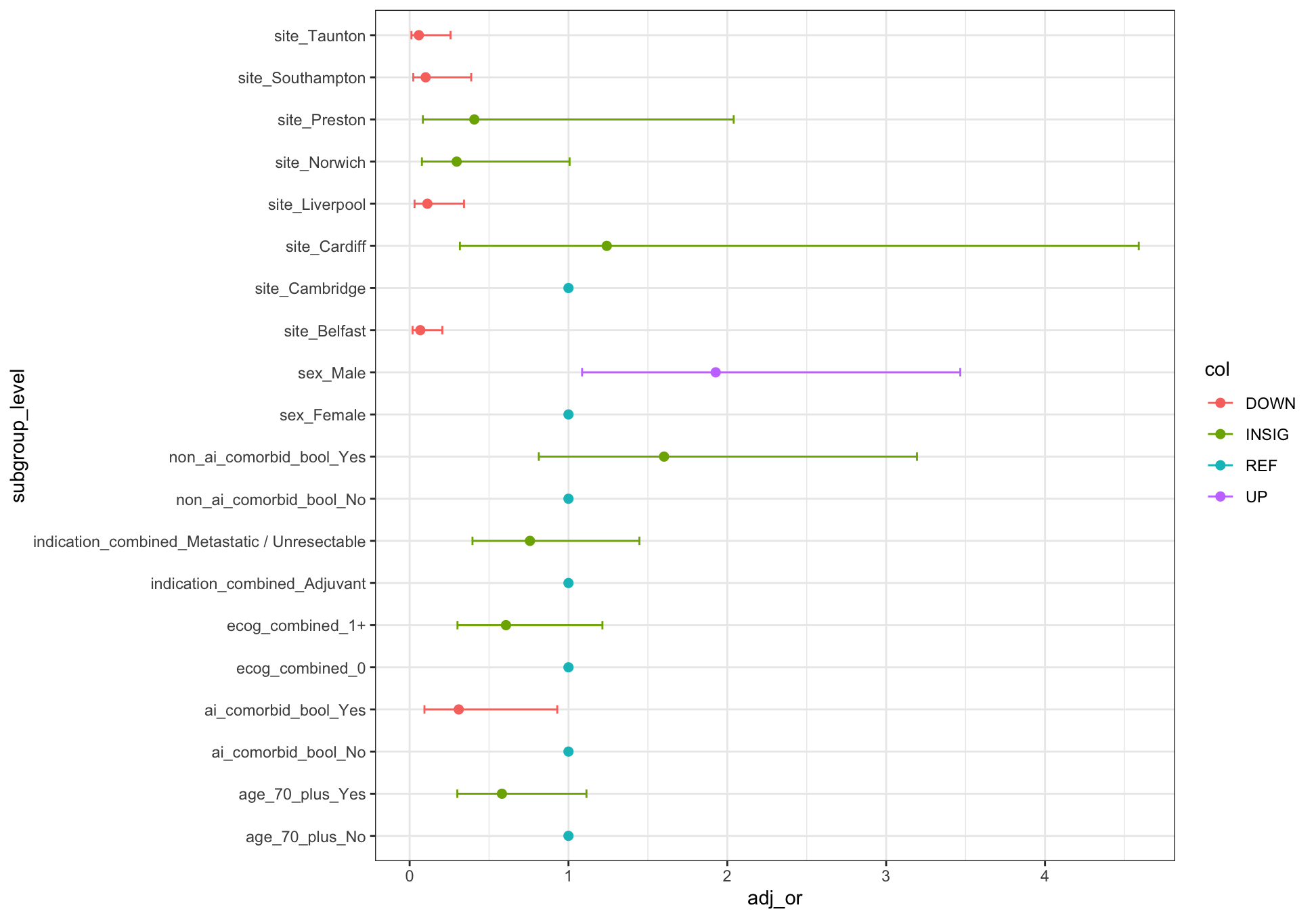
> dput(fig2b_data)
structure(list(subgroup = c("sex", "sex", "ai_comorbid_bool",
"ai_comorbid_bool", "non_ai_comorbid_bool", "non_ai_comorbid_bool",
"age_70_plus", "age_70_plus", "ecog_combined", "ecog_combined",
"indication_combined", "indication_combined", "site", "site",
"site", "site", "site", "site", "site", "site"), level = c("Female",
"Male", "No", "Yes", "No", "Yes", "No", "Yes", "0", "1 ", "Adjuvant",
"Metastatic / Unresectable", "Cambridge", "Belfast", "Cardiff",
"Liverpool", "Norwich", "Preston", "Southampton", "Taunton"),
subgroup_level = c("sex_Female", "sex_Male", "ai_comorbid_bool_No",
"ai_comorbid_bool_Yes", "non_ai_comorbid_bool_No", "non_ai_comorbid_bool_Yes",
"age_70_plus_No", "age_70_plus_Yes", "ecog_combined_0", "ecog_combined_1 ",
"indication_combined_Adjuvant", "indication_combined_Metastatic / Unresectable",
"site_Cambridge", "site_Belfast", "site_Cardiff", "site_Liverpool",
"site_Norwich", "site_Preston", "site_Southampton", "site_Taunton"
), ref = c(TRUE, FALSE, TRUE, FALSE, TRUE, FALSE, TRUE, FALSE,
TRUE, FALSE, TRUE, FALSE, TRUE, FALSE, FALSE, FALSE, FALSE,
FALSE, FALSE, FALSE), adj_or = c(1, 1.92697788983048, 1,
0.309313271153888, 1, 1.60176654927755, 1, 0.581067651194834,
1, 0.606677244239784, 1, 0.757510322046024, 1, 0.0671548910659019,
1.24115412701041, 0.111740502056371, 0.296334401152569, 0.407313416513578,
0.100703132319318, 0.0580853387590806), ci_low = c(NA, 1.08574689964253,
NA, 0.0933004210866726, NA, 0.813446935851162, NA, 0.300096568750007,
NA, 0.301300997438692, NA, 0.395638695943013, NA, 0.0184879397812241,
0.316512222510664, 0.0310182213975059, 0.0774035454553755,
0.0834303368267395, 0.0228743220824828, 0.011193138928203
), ci_high = c(NA, 3.4667621174982, NA, 0.929482385449043,
NA, 3.1938659749789, NA, 1.11325241104074, NA, 1.21374279615277,
NA, 1.44670881667103, NA, 0.205952672316014, 4.59055508109202,
0.342443550375257, 1.00710088916867, 2.04034216674928, 0.387728614421501,
0.257636420370032), p = c(NA, 0.0263295963311719, NA, 0.0432646112707497,
NA, 0.175314541854903, NA, 0.103298047943536, NA, 0.158264479732785,
NA, 0.399589361570504, NA, 8.78601713425597e-06, 0.747238599523183,
0.000291277241946869, 0.0597081504970594, 0.260985385401162,
0.00132018341690714, 0.000328378914869459), sig = c(NA, TRUE,
NA, TRUE, NA, FALSE, NA, FALSE, NA, FALSE, NA, FALSE, NA,
TRUE, FALSE, TRUE, FALSE, FALSE, TRUE, TRUE), col = c("REF",
"UP", "REF", "DOWN", "REF", "INSIG", "REF", "INSIG", "REF",
"INSIG", "REF", "INSIG", "REF", "DOWN", "INSIG", "DOWN",
"INSIG", "INSIG", "DOWN", "DOWN")), row.names = c(NA, -20L
), class = "data.frame")
I'd like to draw a forest plot, but where each level is grouped by subgroup. How can I do this?
I've tried this so far to get my plot, but struggling with grouping the levels:
........................................................................................................................................................................................................
# plot
ggplot(data = fig2b_data, aes(x = adj_or, y = subgroup_level))
geom_errorbarh(aes(xmax = ci_high, xmin = ci_low, color = col), size = .5, height = .2)
geom_point(aes(color = col), size = 2)
theme_bw()
I would like:
_________________________
Sex
Male (ref) x
Female |----x-----|
_______________________________________________________________
AI comorbid
No (ref) x
Yes |----x-----|
_______________________________________________________________
Site
Cambridge(ref) x
Preston |----x-----|
Southampton |----x-----|
Belfast |----x-----|
__________________________________1____________________________
CodePudding user response:
That was quite of a workaround. The adjusted hight for the facet_wrap() height is borrowed from