So I am running multiple cochrane orcutt regressions, which is no problem. I then display the output of these regressions using modelsummary(). Still no problems up to this point.
However, when I then try to compare the models using modelplot(), there are no confidence intervals computed in the cochrane orcutt model (class "orcutt") and I thus get the following error:
Error in eval(parse(text = text, keep.source = FALSE), envir) : object 'conf.low' not found
I know what the problem here is - there are just no confidence interval "parts" computed by the cochrane.orcutt() command. A partial solution is also obvious - I can just calculate the confidence intervals using the point estimates/coefficients and the standard errors (which are of course included in the model by default).
However my problem arises when I want to use these confidence interval values in modelplot(), because they are not "in" the model object. In my ignorance, I attempted the following to try and create the lower bound of a confidence interval, using mutate():
model %>%
mutate(`conf.low`=`coefficients`-1.96*`std.error`)
I hope this conveys my problem well enough, thank you for reading.
CodePudding user response:
As noted in the comments, mutate is a function from the dplyr package which is intended to work on data frames, and not on model objects or on modelsummary tables.
Also, please note that I can’t diagnose your problem properly because you did not supply a MINIMAL REPRODUCIBLE EXAMPLE and you don’t event tell us which function you used to estimate the model.
One way to do the Cochrane-Orcutt in R is to use the orcutt package:
library(orcutt)
data(icecream, package = "orcutt")
lm <- lm(cons ~ price income temp, data = icecream)
coch <- cochrane.orcutt(lm)
Behind the scenes, the modelsummary package uses the tidy function from the broom package to extract estimates from model objects:
library(broom)
tidy(coch)
#> # A tibble: 4 × 5
#> term estimate std.error statistic p.value
#> <chr> <dbl> <dbl> <dbl> <dbl>
#> 1 (Intercept) 0.157 0.290 0.543 0.592
#> 2 price -0.892 0.811 -1.10 0.282
#> 3 income 0.00320 0.00155 2.07 0.0488
#> 4 temp 0.00356 0.000555 6.42 0.00000102
From the output above, you see that broom does NOT extract a confidence interval. This could explain why modelsummary can’t print your table/plot.
One alternative option is to instruct modelsummary to use the parameters package to extract estimates instead of broom. This can be achieved by setting a global option:
library(modelsummary)
options(modelsummary_get = "parameters")
modelsummary(coch, statistic = "conf.int", output = "markdown")
| Model 1 | |
|---|---|
| (Intercept) | 0.157 |
| [-0.439, 0.754] | |
| price | -0.892 |
| [-2.562, 0.778] | |
| income | 0.003 |
| [0.000, 0.006] | |
| temp | 0.004 |
| [0.002, 0.005] | |
| Num.Obs. | 30 |
| R2 | 0.649 |
| R2 Adj. | 0.607 |
| rho | 0.401 |
| number.interaction | 11.000 |
| dw.original | 1.021 |
| p.value.original | 0.000 |
| dw.transformed | 1.549 |
| p.value.transformed | 0.051 |
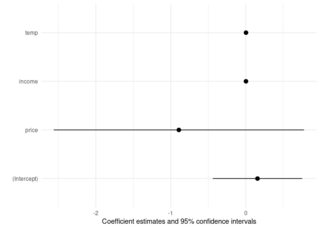
And modelplot now works too:
modelplot(coch)
Yet another alternative would be to use the broom default, but to customize the output using your own tidy_custom.orcutt method. This is a bit more involved, but you’ll find detailed instruction on the modelsummary website: https://vincentarelbundock.github.io/modelsummary/articles/modelsummary.html#adding-new-information-to-existing-models
