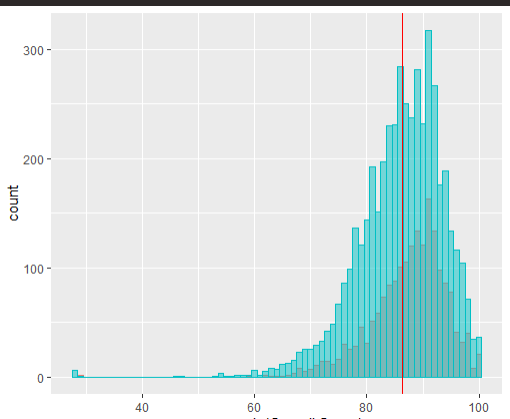
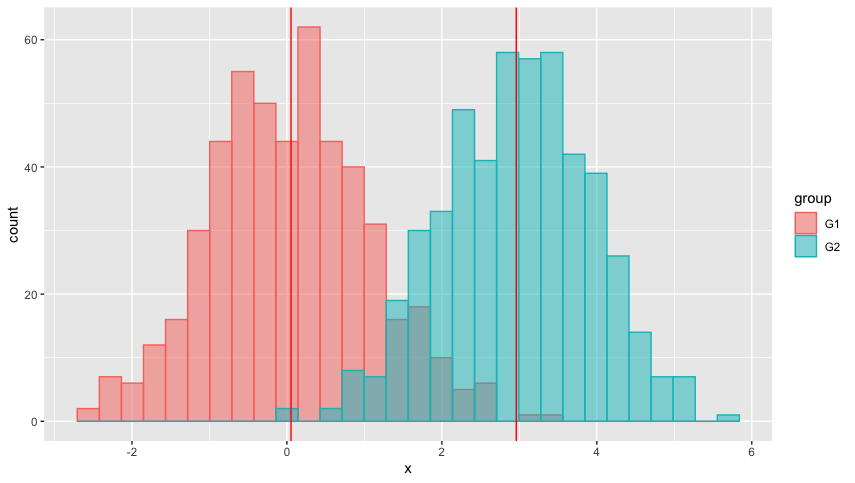
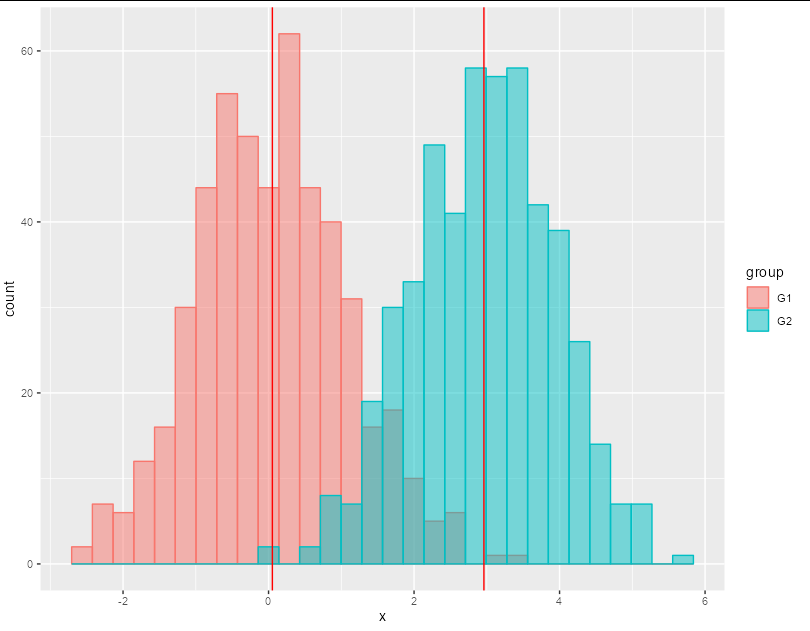
Do you have any suggestions?
CodePudding user response:
In addition to the previous suggestion, you can also use separately stored group means, i. e. two instead of nrow=1000 highly redundant values:
## a 'tidy' (of several valid ways for groupwise calculation):
group_means <- df %>%
group_by(group) %>%
summarise(group_means = mean(x, na.rm = TRUE)) %>%
pull(group_means)
## ... ggplot code ...
geom_vline(xintercept = group_means)
CodePudding user response:
I would compute the mean into the dataframe:
library(ggplot2)
library(dplyr)
df %>%
group_by(group) %>%
mutate(mean_x = mean(x))
output is:
# A tibble: 1,000 × 3
# Groups: group [2]
x group mean_x
<dbl> <chr> <dbl>
1 -0.962 G1 0.0525
2 -0.293 G1 0.0525
3 0.259 G1 0.0525
4 -1.15 G1 0.0525
5 0.196 G1 0.0525
6 0.0301 G1 0.0525
7 0.0854 G1 0.0525
8 1.12 G1 0.0525
9 -1.22 G1 0.0525
10 1.27 G1 0.0525
# … with 990 more rows
So do:
library(ggplot2)
library(dplyr)
df %>%
group_by(group) %>%
mutate(mean_x = mean(x)) %>%
ggplot(aes(x, fill = group, colour = group))
geom_histogram(alpha = 0.5, position = "identity")
geom_vline(aes(xintercept = mean_x), col = "red")
Output is:
CodePudding user response:
A straightforward method without precomputation would be:
ggplot(df, aes(x = x, fill = group, colour = group))
geom_histogram(alpha = 0.5, position = "identity")
geom_vline(xintercept = tapply(df$x, df$group, mean), col = "red")